|
chr22_-_44258210
|
209.719
|
NM_014351
|
SULT4A1
|
sulfotransferase family 4A, member 1
|
|
chr20_+_10199455
|
97.157
|
NM_003081
NM_130811
|
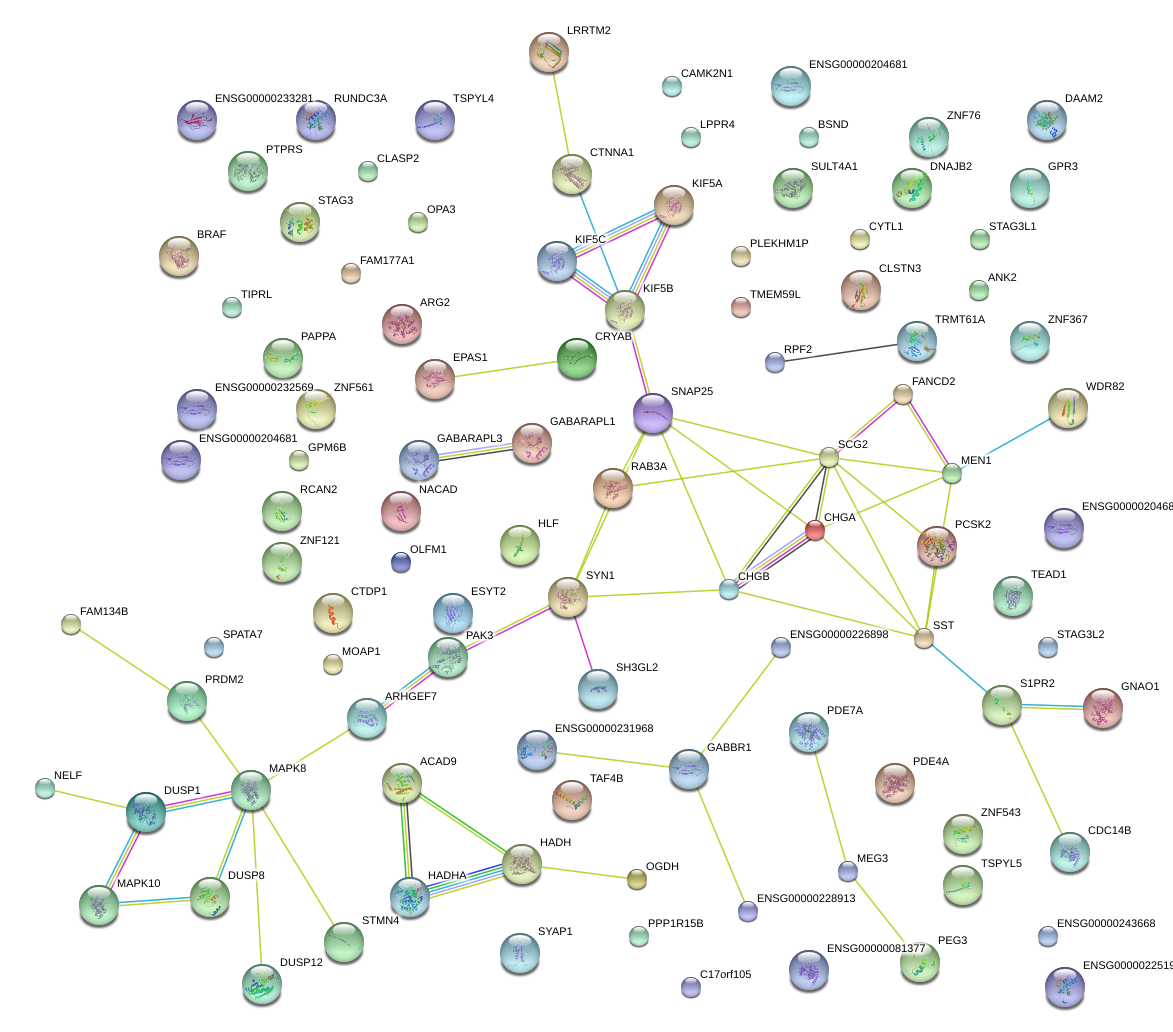
SNAP25
|
synaptosomal-associated protein, 25kDa
|
|
chr19_-_9695166
|
89.120
|
NM_001008727
|
ZNF121
|
zinc finger protein 121
|
|
chr2_+_149632772
|
81.097
|
NM_004522
|
KIF5C
|
kinesin family member 5C
|
|
chr7_+_44646180
|
80.995
|
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide)
|
|
chrX_-_13835187
|
79.424
|
|
GPM6B
|
glycoprotein M6B
|
|
chr7_-_158622184
|
77.849
|
|
ESYT2
|
extended synaptotagmin-like protein 2
|
|
chrX_-_13956644
|
69.721
|
NM_001001994
|
GPM6B
|
glycoprotein M6B
|
|
chr19_-_9731883
|
68.716
|
NM_152289
|
ZNF561
|
zinc finger protein 561
|
|
chr3_-_33759684
|
67.927
|
NM_015097
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr5_-_138210992
|
66.609
|
|
CTNNA1
LRRTM2
|
catenin (cadherin-associated protein), alpha 1, 102kDa
leucine rich repeat transmembrane neuronal 2
|
|
chr8_-_27115935
|
63.630
|
|
STMN4
|
stathmin-like 4
|
|
chr19_+_57831824
|
63.564
|
NM_213598
|
ZNF543
|
zinc finger protein 543
|
|
chr11_-_111782447
|
61.763
|
NM_001885
|
CRYAB
|
crystallin, alpha B
|
|
chrX_-_13835213
|
60.849
|
|
GPM6B
|
glycoprotein M6B
|
|
chr1_-_713985
|
60.475
|
|
LOC100288069
|
general transcription factor IIi pseudogene
|
|
chrX_-_13956530
|
59.774
|
|
GPM6B
|
glycoprotein M6B
|
|
chr12_-_53448167
|
58.956
|
|
LOC283335
|
uncharacterized LOC283335
|
|
chr2_+_149632836
|
58.368
|
|
KIF5C
|
kinesin family member 5C
|
|
chr14_+_101292483
|
57.691
|
|
|
|
|
chr3_+_10028579
|
56.006
|
|
LOC442075
|
uncharacterized LOC442075
|
|
chr14_+_101292451
|
55.440
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr20_+_17207669
|
54.737
|
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2
|
|
chr8_-_27115897
|
54.598
|
NM_030795
|
STMN4
|
stathmin-like 4
|
|
chr1_-_20812727
|
53.767
|
NM_018584
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr2_-_224467095
|
53.657
|
|
SCG2
|
secretogranin II
|
|
chr14_+_93389444
|
53.221
|
NM_001275
|
CHGA
|
chromogranin A (parathyroid secretory protein 1)
|
|
chr14_+_68086570
|
52.760
|
NM_001172
|
ARG2
|
arginase, type II
|
|
chr9_-_140353785
|
52.739
|
NM_001130969
NM_001130970
NM_001130971
NM_001178064
NM_015537
|
NELF
|
nasal embryonic LHRH factor
|
|
chr19_-_18314731
|
52.646
|
NM_002866
|
RAB3A
|
RAB3A, member RAS oncogene family
|
|
chr17_-_62833225
|
52.449
|
|
PLEKHM1P
|
pleckstrin homology domain containing, family M (with RUN domain) member 1 pseudogene
|
|
chr11_+_12695851
|
51.801
|
NM_021961
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor)
|
|
chr5_-_138211054
|
51.675
|
NM_015564
|
LRRTM2
|
leucine rich repeat transmembrane neuronal 2
|
|
chr6_+_39760148
|
51.609
|
NM_001201427
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chr9_-_99382073
|
51.498
|
NM_003671
NM_033331
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr3_-_187388105
|
51.385
|
NM_001048
|
SST
|
somatostatin
|
|
chr19_-_10341947
|
51.008
|
NM_004230
|
S1PR2
|
sphingosine-1-phosphate receptor 2
|
|
chr9_-_140353748
|
50.270
|
|
NELF
|
nasal embryonic LHRH factor
|
|
chrX_-_47479229
|
50.096
|
NM_006950
NM_133499
|
SYN1
|
synapsin I
|
|
chr6_-_29595746
|
49.690
|
|
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1
|
|
chr2_-_224467049
|
49.397
|
|
SCG2
|
secretogranin II
|
|
chr19_+_18723666
|
49.074
|
NM_012109
|
TMEM59L
|
transmembrane protein 59-like
|
|
chr11_-_1593149
|
48.690
|
NM_004420
|
DUSP8
|
dual specificity phosphatase 8
|
|
chr3_-_10067989
|
48.617
|
|
CIDECP
|
cell death-inducing DFFA-like effector c pseudogene
|
|
chr14_+_88851987
|
47.675
|
NM_001040428
NM_018418
|
SPATA7
|
spermatogenesis associated 7
|
|
chrX_-_13835012
|
47.648
|
|
GPM6B
|
glycoprotein M6B
|
|
chr19_-_5340700
|
47.294
|
NM_002850
NM_130853
NM_130854
NM_130855
|
PTPRS
|
protein tyrosine phosphatase, receptor type, S
|
|
chr20_+_5891973
|
47.159
|
NM_001819
|
CHGB
|
chromogranin B (secretogranin 1)
|
|
chr17_+_53342356
|
46.387
|
|
HLF
|
hepatic leukemia factor
|
|
chrX_-_13956445
|
46.086
|
|
|
|
|
chr7_-_74306672
|
45.821
|
|
STAG3L2
|
stromal antigen 3-like 2
|
|
chr1_+_99730067
|
45.317
|
|
LPPR4
|
lipid phosphate phosphatase-related protein type 4
|
|
chr16_+_56225301
|
45.255
|
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O
|
|
chr14_+_93389491
|
44.786
|
|
CHGA
|
chromogranin A (parathyroid secretory protein 1)
|
|
chr14_-_93651242
|
44.516
|
NM_022151
|
MOAP1
|
modulator of apoptosis 1
|
|
chr19_-_57352055
|
44.423
|
NM_001146326
NM_001146327
NM_015363
NM_001146184
NM_001146185
NM_001146187
NM_006210
|
ZIM2
PEG3
|
zinc finger, imprinted 2
paternally expressed 3
|
|
chr8_-_66753968
|
43.853
|
NM_001242318
|
PDE7A
|
phosphodiesterase 7A
|
|
chr6_+_111303240
|
43.427
|
NM_032194
|
RPF2
|
ribosome production factor 2 homolog (S. cerevisiae)
|
|
chr9_+_17578952
|
42.890
|
NM_003026
|
SH3GL2
|
SH3-domain GRB2-like 2
|
|
chr9_+_137967088
|
42.716
|
NM_014279
NM_006334
|
OLFM1
|
olfactomedin 1
|
|
chr3_-_33759540
|
42.627
|
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr2_-_224467142
|
42.268
|
NM_003469
|
SCG2
|
secretogranin II
|
|
chr20_+_17207680
|
41.892
|
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2
|
|
chr1_+_55464605
|
41.586
|
NM_057176
|
BSND
|
Bartter syndrome, infantile, with sensorineural deafness (Barttin)
|
|
chr3_+_10068123
|
40.810
|
|
FANCD2
|
Fanconi anemia, complementation group D2
|
|
chr20_+_17207596
|
40.554
|
NM_001201529
NM_002594
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2
|
|
chr3_-_8543327
|
40.190
|
|
LOC100288428
|
uncharacterized LOC100288428
|
|
chr14_+_88852065
|
38.956
|
|
SPATA7
|
spermatogenesis associated 7
|
|
chr19_-_46088021
|
38.954
|
|
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia)
|
|
chr3_+_128598471
|
38.633
|
|
ACAD9
|
acyl-CoA dehydrogenase family, member 9
|
|
chr17_+_42385780
|
38.562
|
NM_001144825
NM_001144826
NM_006695
|
RUNDC3A
|
RUN domain containing 3A
|
|
chrX_-_13835313
|
38.515
|
NM_001001995
NM_001001996
NM_005278
|
GPM6B
|
glycoprotein M6B
|
|
chr1_+_14026685
|
38.325
|
NM_001135610
|
PRDM2
|
PR domain containing 2, with ZNF domain
|
|
chr1_-_204380903
|
37.488
|
NM_032833
|
PPP1R15B
|
protein phosphatase 1, regulatory subunit 15B
|
|
chr19_-_46088074
|
37.406
|
|
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia)
|
|
chr12_+_10365415
|
36.298
|
NM_031412
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1
|
|
chr3_-_52312257
|
36.224
|
|
WDR82
|
WD repeat domain 82
|
|
chr19_+_10527448
|
35.746
|
NM_001243121
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr6_+_35227234
|
35.478
|
|
ZNF76
|
zinc finger protein 76
|
|
chr7_-_45128492
|
35.477
|
NM_001146334
|
NACAD
|
NAC alpha domain containing
|
|
chr1_+_27719151
|
35.460
|
NM_005281
|
GPR3
|
G protein-coupled receptor 3
|
|
chr11_-_65337743
|
35.417
|
|
LOC254100
|
uncharacterized LOC254100
|
|
chrX_+_110339374
|
35.000
|
NM_002578
|
PAK3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr7_-_140624280
|
34.739
|
NM_004333
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B1
|
|
chr18_+_23806385
|
34.230
|
NM_005640
|
TAF4B
|
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa
|
|
chr14_-_103995336
|
34.032
|
|
|
|
|
chr9_+_137967401
|
33.520
|
|
OLFM1
|
olfactomedin 1
|
|
chr2_+_149633173
|
33.484
|
|
|
|
|
chr4_+_113739203
|
33.449
|
NM_001127493
|
ANK2
|
ankyrin 2, neuronal
|
|
chr2_+_220144024
|
33.338
|
NM_001039550
NM_006736
|
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2
|
|
chrX_+_16737732
|
32.911
|
|
SYAP1
|
synapse associated protein 1
|
|
chr12_+_7282963
|
32.382
|
NM_014718
|
CLSTN3
|
calsyntenin 3
|
|
chr14_+_35515347
|
32.328
|
NM_173607
|
FAM177A1
|
family with sequence similarity 177, member A1
|
|
chr14_+_68086655
|
32.248
|
|
ARG2
|
arginase, type II
|
|
chr9_+_137967366
|
31.833
|
|
OLFM1
|
olfactomedin 1
|
|
chr17_+_41857802
|
31.587
|
NM_001136483
|
C17orf105
|
chromosome 17 open reading frame 105
|
|
chr5_-_16617085
|
31.461
|
NM_001034850
|
FAM134B
|
family with sequence similarity 134, member B
|
|
chr5_-_172198160
|
30.894
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chr8_-_98290072
|
30.613
|
|
TSPYL5
|
TSPY-like 5
|
|
chr14_+_103995535
|
30.353
|
|
TRMT61A
|
tRNA methyltransferase 61 homolog A (S. cerevisiae)
|
|
chr6_-_29595934
|
30.259
|
NM_021903
|
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1
|
|
chr4_+_108911008
|
30.248
|
|
HADH
|
hydroxyacyl-CoA dehydrogenase
|
|
chr6_-_46293628
|
30.184
|
NM_005822
|
RCAN2
|
regulator of calcineurin 2
|
|
chr18_+_77439798
|
30.048
|
NM_004715
NM_048368
|
CTDP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1
|
|
chr1_+_168148215
|
29.983
|
|
TIPRL
|
TIP41, TOR signaling pathway regulator-like (S. cerevisiae)
|
|
chr17_-_62833264
|
29.687
|
|
PLEKHM1P
|
pleckstrin homology domain containing, family M (with RUN domain) member 1 pseudogene
|
|
chr4_-_87281177
|
29.427
|
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chrX_+_16737661
|
28.724
|
NM_032796
|
SYAP1
|
synapse associated protein 1
|
|
chr4_+_668250
|
28.605
|
|
MYL5
|
myosin, light chain 5, regulatory
|
|
chr17_+_53342320
|
28.501
|
NM_002126
|
HLF
|
hepatic leukemia factor
|
|
chr16_+_56225250
|
28.126
|
NM_020988
NM_138736
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O
|
|
chr1_-_235812994
|
27.761
|
NM_001098722
|
GNG4
|
guanine nucleotide binding protein (G protein), gamma 4
|
|
chr6_-_91006580
|
27.631
|
NM_001170794
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr7_-_8301681
|
27.584
|
NM_022307
|
ICA1
|
islet cell autoantigen 1, 69kDa
|
|
chr6_-_46293404
|
27.548
|
|
RCAN2
|
regulator of calcineurin 2
|
|
chr4_+_668312
|
27.394
|
|
MYL5
|
myosin, light chain 5, regulatory
|
|
chr11_-_119234859
|
27.363
|
NM_171997
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr2_-_198175324
|
27.242
|
|
ANKRD44
|
ankyrin repeat domain 44
|
|
chr2_-_26700909
|
27.082
|
NM_004802
NM_194322
NM_194323
|
OTOF
|
otoferlin
|
|
chr1_+_29562947
|
27.036
|
NM_001195001
NM_005704
NM_133177
NM_133178
|
PTPRU
|
protein tyrosine phosphatase, receptor type, U
|
|
chr6_-_91006460
|
26.992
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr2_+_11052062
|
26.924
|
NM_002236
|
KCNF1
|
potassium voltage-gated channel, subfamily F, member 1
|
|
chr19_+_57791794
|
26.760
|
NM_006635
|
ZNF460
|
zinc finger protein 460
|
|
chr14_+_68086633
|
26.648
|
|
ARG2
|
arginase, type II
|
|
chr10_-_73848263
|
26.646
|
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2
|
|
chr13_-_29069215
|
26.617
|
NM_001159920
NM_001160030
NM_001160031
NM_002019
|
FLT1
|
fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor)
|
|
chr12_-_16760935
|
26.455
|
NM_001001395
NM_001243609
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr18_+_77439843
|
26.427
|
|
CTDP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1
|
|
chr13_-_41495804
|
26.167
|
|
LOC100616668
SUGT1P3
|
TPTE2 pseudogene
suppressor of G2 allele of SKP1 (S. cerevisiae) pseudogene 3
|
|
chr8_-_102803178
|
26.010
|
|
NCALD
|
neurocalcin delta
|
|
chr15_+_91769329
|
25.960
|
|
SV2B
|
synaptic vesicle glycoprotein 2B
|
|
chr8_-_102803196
|
25.829
|
|
NCALD
|
neurocalcin delta
|
|
chr17_+_39845142
|
25.811
|
|
EIF1
|
eukaryotic translation initiation factor 1
|
|
chr19_+_45504686
|
25.704
|
NM_006509
|
RELB
|
v-rel reticuloendotheliosis viral oncogene homolog B
|
|
chr17_+_29421956
|
25.623
|
|
NF1
|
neurofibromin 1
|
|
chr2_-_192015706
|
25.458
|
|
STAT4
|
signal transducer and activator of transcription 4
|
|
chr19_-_36246411
|
25.398
|
|
HSPB6
|
heat shock protein, alpha-crystallin-related, B6
|
|
chr1_-_35325335
|
25.299
|
NM_001164825
NM_138428
|
C1orf212
|
chromosome 1 open reading frame 212
|
|
chr2_+_105953815
|
25.260
|
|
C2orf49
|
chromosome 2 open reading frame 49
|
|
chr10_+_14920781
|
25.249
|
NM_001193424
NM_001193426
NM_001193425
NM_001193427
NM_024670
|
SUV39H2
|
suppressor of variegation 3-9 homolog 2 (Drosophila)
|
|
chr9_+_105757592
|
25.098
|
NM_001340
|
CYLC2
|
cylicin, basic protein of sperm head cytoskeleton 2
|
|
chr15_+_68871285
|
24.959
|
NM_006091
|
CORO2B
|
coronin, actin binding protein, 2B
|
|
chr11_-_62389571
|
24.853
|
NM_012200
|
B3GAT3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I)
|
|
chr12_-_13256572
|
24.833
|
NM_001080554
NM_001080555
NM_001206842
|
GSG1
|
germ cell associated 1
|
|
chr14_+_103995473
|
24.753
|
NM_152307
|
TRMT61A
|
tRNA methyltransferase 61 homolog A (S. cerevisiae)
|
|
chr14_-_103988829
|
24.646
|
|
CKB
|
creatine kinase, brain
|
|
chr2_-_216300770
|
24.635
|
NM_002026
NM_054034
NM_212474
NM_212476
NM_212478
NM_212482
|
FN1
|
fibronectin 1
|
|
chr5_+_71502700
|
24.460
|
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr7_+_74988447
|
24.393
|
|
STAG3L3
STAG3L2
STAG3L1
|
stromal antigen 3-like 3
stromal antigen 3-like 2
stromal antigen 3-like 1
|
|
chr6_-_121655606
|
24.354
|
NM_152730
|
C6orf170
|
chromosome 6 open reading frame 170
|
|
chr21_-_45079327
|
24.112
|
NM_007031
|
HSF2BP
|
heat shock transcription factor 2 binding protein
|
|
chr17_+_79885567
|
24.094
|
|
LOC92659
|
uncharacterized LOC92659
|
|
chr16_+_67694795
|
23.908
|
NM_001037281
NM_016948
|
PARD6A
|
par-6 partitioning defective 6 homolog alpha (C. elegans)
|
|
chr11_+_117070039
|
23.871
|
NM_001001522
NM_003186
|
TAGLN
|
transgelin
|
|
chr14_+_104095491
|
23.842
|
NM_001130107
NM_005552
NM_182923
|
KLC1
|
kinesin light chain 1
|
|
chr14_-_36278335
|
23.834
|
NM_014990
NM_194301
|
RALGAPA1
|
Ral GTPase activating protein, alpha subunit 1 (catalytic)
|
|
chr17_+_39845141
|
23.822
|
|
EIF1
|
eukaryotic translation initiation factor 1
|
|
chr12_+_107168336
|
23.817
|
NM_018157
|
RIC8B
|
resistance to inhibitors of cholinesterase 8 homolog B (C. elegans)
|
|
chr22_-_20104745
|
23.760
|
NM_022727
NM_182984
|
TRMT2A
|
TRM2 tRNA methyltransferase 2 homolog A (S. cerevisiae)
|
|
chr5_-_112770537
|
23.579
|
NM_032028
|
TSSK1B
|
testis-specific serine kinase 1B
|
|
chr16_+_29912146
|
23.376
|
NM_181718
|
ASPHD1
|
aspartate beta-hydroxylase domain containing 1
|
|
chr22_-_20104695
|
23.297
|
|
TRMT2A
|
TRM2 tRNA methyltransferase 2 homolog A (S. cerevisiae)
|
|
chrX_-_92928566
|
23.249
|
NM_004538
|
NAP1L3
|
nucleosome assembly protein 1-like 3
|
|
chr5_-_95768660
|
23.154
|
NM_000439
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|
|
chr2_-_220408264
|
23.060
|
NM_024536
|
CHPF
|
chondroitin polymerizing factor
|
|
chr8_-_98290171
|
23.052
|
NM_033512
|
TSPYL5
|
TSPY-like 5
|
|
chr5_-_132948222
|
23.046
|
NM_015082
|
FSTL4
|
follistatin-like 4
|
|
chr8_-_18666295
|
23.018
|
NM_206909
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr7_-_8301767
|
22.966
|
NM_001136020
|
ICA1
|
islet cell autoantigen 1, 69kDa
|
|
chr17_+_41476351
|
22.519
|
NM_001661
|
ARL4D
|
ADP-ribosylation factor-like 4D
|
|
chr1_+_2005083
|
22.413
|
NM_001033581
|
PRKCZ
|
protein kinase C, zeta
|
|
chr10_+_64133915
|
22.364
|
NM_014951
NM_199450
NM_199451
|
ZNF365
|
zinc finger protein 365
|
|
chr4_-_4249928
|
22.293
|
NM_032927
|
TMEM128
|
transmembrane protein 128
|
|
chr3_+_184053682
|
22.290
|
NM_001171093
|
FAM131A
|
family with sequence similarity 131, member A
|
|
chr16_+_22825806
|
22.288
|
NM_006043
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2
|
|
chr17_-_43568081
|
22.113
|
|
PLEKHM1
|
pleckstrin homology domain containing, family M (with RUN domain) member 1
|
|
chr1_+_110527418
|
22.096
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr7_+_66767575
|
22.065
|
|
STAG3L4
|
stromal antigen 3-like 4
|
|
chr1_+_119683006
|
22.041
|
|
|
|
|
chr16_-_90038869
|
21.977
|
|
CENPBD1
|
CENPB DNA-binding domains containing 1
|
|
chr5_-_133706717
|
21.971
|
|
CDKL3
|
cyclin-dependent kinase-like 3
|
|
chr2_-_220174058
|
21.960
|
|
PTPRN
|
protein tyrosine phosphatase, receptor type, N
|
|
chr3_+_50297644
|
21.945
|
|
|
|
|
chr6_-_3157696
|
21.866
|
NM_001069
|
TUBB2A
|
tubulin, beta 2A class IIa
|
|
chr8_+_27631902
|
21.823
|
NM_001017420
|
ESCO2
|
establishment of cohesion 1 homolog 2 (S. cerevisiae)
|
|
chr1_+_36023389
|
21.756
|
NM_001014839
NM_001014841
NM_014284
|
NCDN
|
neurochondrin
|
|
chr16_+_22825481
|
21.671
|
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2
|
|
chr8_-_21966812
|
21.352
|
NM_024815
|
NUDT18
|
nudix (nucleoside diphosphate linked moiety X)-type motif 18
|
|
chr5_-_95297424
|
21.290
|
|
ELL2
|
elongation factor, RNA polymerase II, 2
|
|
chr19_-_40919270
|
21.266
|
NM_020956
NM_181882
|
PRX
|
periaxin
|
|
chr9_-_130477918
|
21.005
|
NM_001002913
|
PTRH1
|
peptidyl-tRNA hydrolase 1 homolog (S. cerevisiae)
|
|
chr4_-_170533536
|
20.861
|
|
NEK1
|
NIMA (never in mitosis gene a)-related kinase 1
|
|
chr11_+_62475165
|
20.853
|
|
GNG3
|
guanine nucleotide binding protein (G protein), gamma 3
|
|
chr6_-_169653922
|
20.706
|
|
THBS2
|
thrombospondin 2
|
|
chr2_+_24150144
|
20.623
|
|
UBXN2A
|
UBX domain protein 2A
|
|
chr17_-_62833246
|
20.548
|
|
PLEKHM1P
|
pleckstrin homology domain containing, family M (with RUN domain) member 1 pseudogene
|
|
chr14_+_76618272
|
20.370
|
|
C14orf118
|
chromosome 14 open reading frame 118
|
|
chr4_+_175204827
|
20.353
|
NM_001040157
|
CEP44
|
centrosomal protein 44kDa
|
|
chr8_+_80523304
|
20.252
|
|
STMN2
|
stathmin-like 2
|
|
chr14_-_93651182
|
20.194
|
|
MOAP1
|
modulator of apoptosis 1
|