|
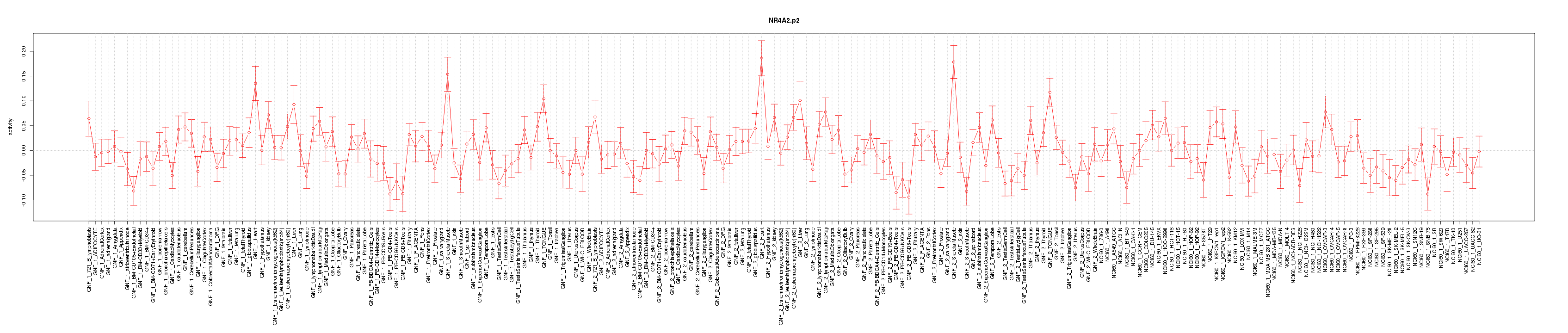
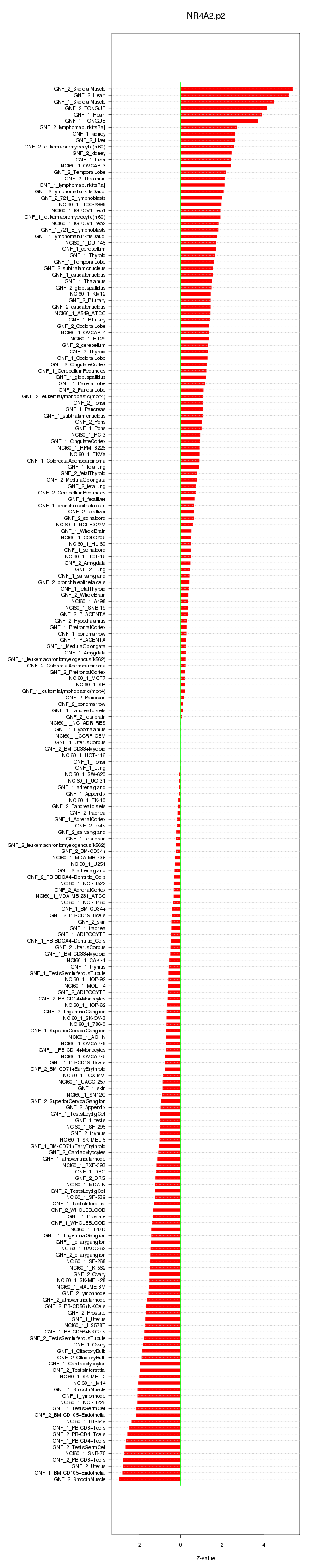
chr20_-_62130390
|
24.008
|
NM_001958
|
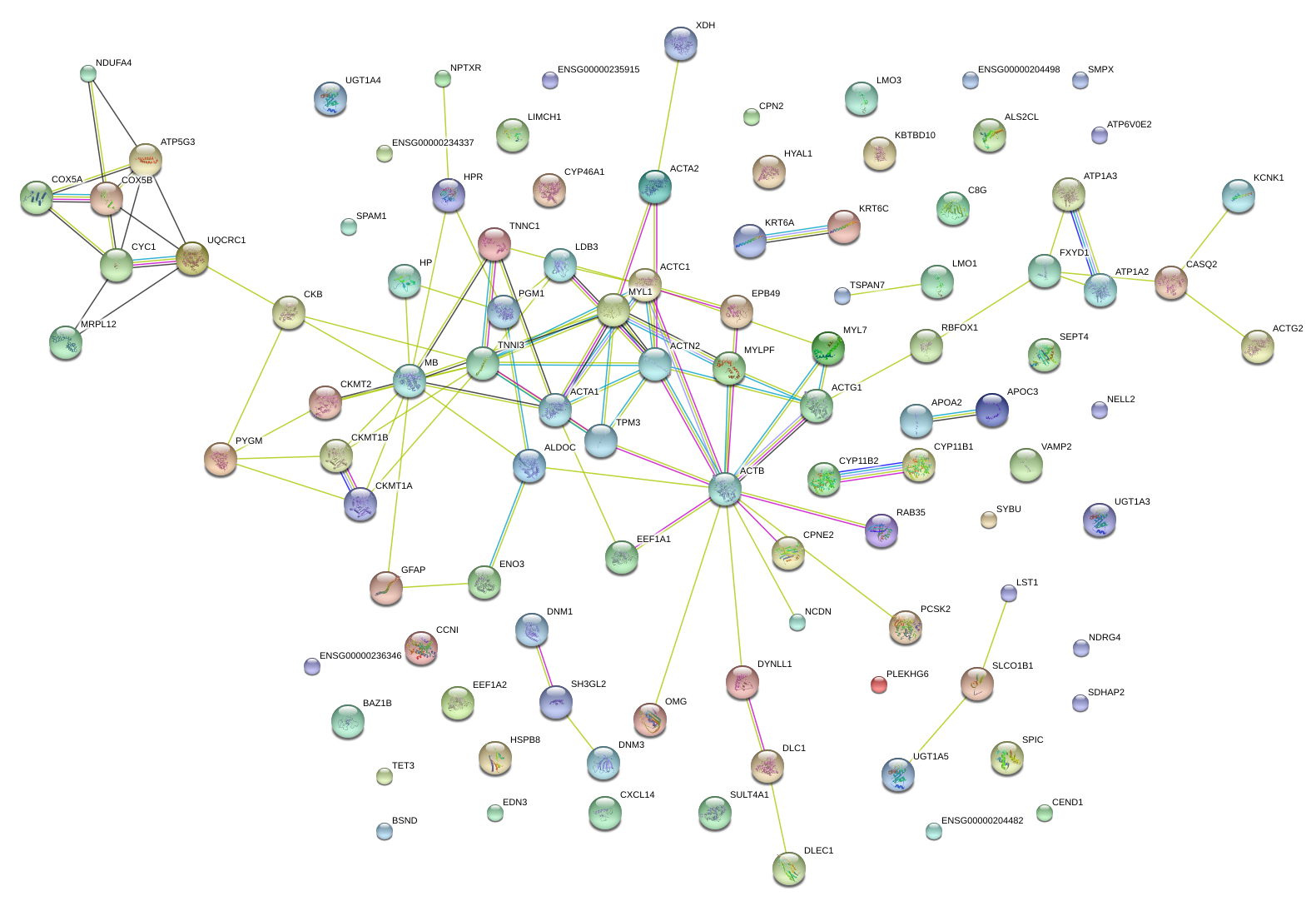
EEF1A2
|
eukaryotic translation elongation factor 1 alpha 2
|
|
chr12_-_16759531
|
22.737
|
NM_018640
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr12_-_16759430
|
20.469
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr12_-_16759939
|
19.086
|
NM_001243613
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr1_+_236849753
|
18.130
|
NM_001103
|
ACTN2
|
actinin, alpha 2
|
|
chr12_-_16759733
|
14.696
|
NM_001243610
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr5_+_80529138
|
14.666
|
NM_001099735
NM_001099736
NM_001825
|
CKMT2
|
creatine kinase, mitochondrial 2 (sarcomeric)
|
|
chr12_-_16760935
|
13.682
|
NM_001001395
NM_001243609
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr12_-_16758839
|
13.641
|
NM_001243612
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr8_+_145149985
|
13.211
|
|
CYC1
|
cytochrome c-1
|
|
chr1_-_229569839
|
13.125
|
NM_001100
|
ACTA1
|
actin, alpha 1, skeletal muscle
|
|
chr11_-_64528186
|
12.903
|
NM_001164716
NM_005609
|
PYGM
|
phosphorylase, glycogen, muscle
|
|
chr22_-_36019363
|
12.569
|
NM_203377
|
MB
|
myoglobin
|
|
chr1_-_154164551
|
12.391
|
NM_152263
|
TPM3
|
tropomyosin 3
|
|
chr17_+_4854383
|
12.274
|
NM_001193503
NM_001976
NM_053013
|
ENO3
|
enolase 3 (beta, muscle)
|
|
chr3_-_47957475
|
11.905
|
|
|
|
|
chr15_+_43885251
|
10.971
|
NM_020990
|
CKMT1B
CKMT1A
|
creatine kinase, mitochondrial 1B
creatine kinase, mitochondrial 1A
|
|
chr1_+_55464605
|
9.970
|
NM_057176
|
BSND
|
Bartter syndrome, infantile, with sensorineural deafness (Barttin)
|
|
chr19_+_35629727
|
9.412
|
NM_021902
|
FXYD1
|
FXYD domain containing ion transport regulator 1
|
|
chrX_+_38420690
|
8.712
|
NM_004615
|
TSPAN7
|
tetraspanin 7
|
|
chr16_+_58534045
|
8.462
|
NM_001242833
NM_001242834
NM_001242835
NM_001242836
|
NDRG4
|
NDRG family member 4
|
|
chr14_-_103988829
|
8.237
|
|
CKB
|
creatine kinase, brain
|
|
chr17_+_79670396
|
8.141
|
NM_002949
|
MRPL12
|
mitochondrial ribosomal protein L12
|
|
chr2_-_176046120
|
8.098
|
NM_001002258
NM_001190329
NM_001689
|
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9)
|
|
chr7_-_10979683
|
7.950
|
|
NDUFA4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4, 9kDa
|
|
chr1_+_160085519
|
7.925
|
NM_000702
|
ATP1A2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide
|
|
chr3_-_46718898
|
7.660
|
NM_182775
|
ALS2CL
|
ALS2 C-terminal like
|
|
chr3_-_50340979
|
7.635
|
NM_007312
NM_033159
NM_153282
NM_153283
NM_153285
|
HYAL1
|
hyaluronoglucosaminidase 1
|
|
chr17_+_79670495
|
7.512
|
|
MRPL12
|
mitochondrial ribosomal protein L12
|
|
chr1_+_171810611
|
7.492
|
NM_001136127
NM_015569
|
DNM3
|
dynamin 3
|
|
chr9_+_130965666
|
7.471
|
|
DNM1
|
dynamin 1
|
|
chr1_+_36023389
|
7.434
|
NM_001014839
NM_001014841
NM_014284
|
NCDN
|
neurochondrin
|
|
chr16_+_72097124
|
7.287
|
NM_020995
|
HPR
|
haptoglobin-related protein
|
|
chr10_+_88428167
|
6.949
|
NM_001171610
|
LDB3
|
LIM domain binding 3
|
|
chr2_+_234621637
|
6.926
|
NM_019078
|
UGT1A5
|
UDP glucuronosyltransferase 1 family, polypeptide A5
|
|
chr12_+_6421816
|
6.870
|
NM_001144857
|
PLEKHG6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6
|
|
chr16_+_6069094
|
6.826
|
NM_001142333
NM_018723
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr22_-_39240016
|
6.757
|
NM_014293
|
NPTXR
|
neuronal pentraxin receptor
|
|
chr2_-_211168315
|
6.727
|
NM_079422
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast
|
|
chr9_+_130965608
|
6.710
|
NM_001005336
NM_004408
|
DNM1
|
dynamin 1
|
|
chr11_+_116700620
|
6.707
|
NM_000040
|
APOC3
|
apolipoprotein C-III
|
|
chr17_-_26903926
|
6.689
|
NM_005165
|
ALDOC
|
aldolase C, fructose-bisphosphate
|
|
chr15_-_75230354
|
6.614
|
|
COX5A
|
cytochrome c oxidase subunit Va
|
|
chr20_+_17207680
|
6.559
|
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2
|
|
chr9_+_130965668
|
6.475
|
|
DNM1
|
dynamin 1
|
|
chr3_-_194072004
|
6.474
|
NM_001080513
|
CPN2
|
carboxypeptidase N, polypeptide 2
|
|
chr1_+_64088886
|
6.282
|
NM_001172818
|
PGM1
|
phosphoglucomutase 1
|
|
chr17_-_42992852
|
6.275
|
NM_001131019
NM_001242376
NM_002055
|
GFAP
|
glial fibrillary acidic protein
|
|
chr11_+_65190272
|
6.245
|
|
|
|
|
chr14_+_100150596
|
6.166
|
NM_006668
|
CYP46A1
|
cytochrome P450, family 46, subfamily A, polypeptide 1
|
|
chr8_+_145149978
|
6.155
|
|
CYC1
|
cytochrome c-1
|
|
chr2_+_74273449
|
6.038
|
NM_144993
|
TET3
|
tet methylcytosine dioxygenase 3
|
|
chr8_+_145149936
|
5.945
|
NM_001916
|
CYC1
|
cytochrome c-1
|
|
chr12_-_120554558
|
5.938
|
|
RAB35
|
RAB35, member RAS oncogene family
|
|
chr3_-_48647052
|
5.937
|
|
UQCRC1
|
ubiquinol-cytochrome c reductase core protein I
|
|
chr3_-_48647085
|
5.919
|
NM_003365
|
UQCRC1
|
ubiquinol-cytochrome c reductase core protein I
|
|
chr22_-_44258210
|
5.895
|
NM_014351
|
SULT4A1
|
sulfotransferase family 4A, member 1
|
|
chr17_-_26903854
|
5.829
|
|
ALDOC
|
aldolase C, fructose-bisphosphate
|
|
chr7_+_149571029
|
5.823
|
|
ATP6V0E2
|
ATPase, H+ transporting V0 subunit e2
|
|
chr7_-_44180911
|
5.821
|
NM_021223
|
MYL7
|
myosin, light chain 7, regulatory
|
|
chr3_+_195384918
|
5.763
|
|
SDHAP1
SDHAP2
|
succinate dehydrogenase complex, subunit A, flavoprotein pseudogene 1
succinate dehydrogenase complex, subunit A, flavoprotein pseudogene 2
|
|
chr1_+_233749749
|
5.649
|
NM_002245
|
KCNK1
|
potassium channel, subfamily K, member 1
|
|
chr4_+_41614911
|
5.646
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chrX_-_21776158
|
5.533
|
NM_014332
|
SMPX
|
small muscle protein, X-linked
|
|
chr2_+_234637772
|
5.476
|
NM_019093
|
UGT1A3
|
UDP glucuronosyltransferase 1 family, polypeptide A3
|
|
chr19_+_35630391
|
5.290
|
NM_005031
|
FXYD1
|
FXYD domain containing ion transport regulator 1
|
|
chr12_-_45307710
|
5.269
|
NM_001145110
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr9_+_17578952
|
5.265
|
NM_003026
|
SH3GL2
|
SH3-domain GRB2-like 2
|
|
chr5_-_134914968
|
5.201
|
NM_004887
|
CXCL14
|
chemokine (C-X-C motif) ligand 14
|
|
chr16_+_30386097
|
5.186
|
NM_013292
|
MYLPF
|
myosin light chain, phosphorylatable, fast skeletal muscle
|
|
chr1_-_161193190
|
5.131
|
|
APOA2
|
apolipoprotein A-II
|
|
chr2_+_170366211
|
5.128
|
NM_006063
|
KBTBD10
|
kelch repeat and BTB (POZ) domain containing 10
|
|
chr7_-_72936592
|
5.079
|
NM_032408
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B
|
|
chr17_-_8066254
|
5.075
|
NM_014232
|
VAMP2
|
vesicle-associated membrane protein 2 (synaptobrevin 2)
|
|
chr2_+_98262497
|
5.050
|
NM_001862
|
COX5B
|
cytochrome c oxidase subunit Vb
|
|
chr2_+_234627423
|
4.963
|
NM_007120
|
UGT1A4
|
UDP glucuronosyltransferase 1 family, polypeptide A4
|
|
chr1_-_116311161
|
4.902
|
|
CASQ2
|
calsequestrin 2 (cardiac muscle)
|
|
chr17_-_56618178
|
4.882
|
NM_001198713
|
SEPT4
|
septin 4
|
|
chr20_+_57875647
|
4.873
|
|
EDN3
|
endothelin 3
|
|
chr3_-_52488030
|
4.871
|
NM_003280
|
TNNC1
|
troponin C type 1 (slow)
|
|
chr17_-_29624249
|
4.864
|
NM_002544
|
OMG
|
oligodendrocyte myelin glycoprotein
|
|
chr16_+_72088507
|
4.855
|
NM_001126102
NM_005143
|
HP
|
haptoglobin
|
|
chr2_-_31637553
|
4.824
|
|
XDH
|
xanthine dehydrogenase
|
|
chr12_-_52886971
|
4.810
|
NM_005554
|
KRT6A
|
keratin 6A
|
|
chr15_-_75230386
|
4.806
|
NM_004255
|
COX5A
|
cytochrome c oxidase subunit Va
|
|
chr12_+_119616594
|
4.789
|
NM_014365
|
HSPB8
|
heat shock 22kDa protein 8
|
|
chr1_-_116311330
|
4.754
|
|
CASQ2
|
calsequestrin 2 (cardiac muscle)
|
|
chr6_+_31554475
|
4.731
|
NM_205837
|
LST1
|
leukocyte specific transcript 1
|
|
chr11_-_790103
|
4.611
|
NM_016564
|
CEND1
|
cell cycle exit and neuronal differentiation 1
|
|
chr9_+_139839697
|
4.609
|
NM_000606
|
C8G
|
complement component 8, gamma polypeptide
|
|
chr1_+_64059479
|
4.600
|
NM_001172819
|
PGM1
|
phosphoglucomutase 1
|
|
chr8_-_110661007
|
4.571
|
NM_001099747
NM_001099748
NM_001099749
NM_001099750
NM_017786
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr8_+_21911062
|
4.553
|
NM_001114137
|
EPB49
|
erythrocyte membrane protein band 4.9 (dematin)
|
|
chr14_-_103989112
|
4.514
|
|
CKB
|
creatine kinase, brain
|
|
chr1_-_182573393
|
4.486
|
NM_002928
|
RGS16
|
regulator of G-protein signaling 16
|
|
chr19_+_54606141
|
4.484
|
NM_004542
|
NDUFA3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3, 9kDa
|
|
chr15_+_91769329
|
4.471
|
|
SV2B
|
synaptic vesicle glycoprotein 2B
|
|
chr17_-_56606710
|
4.462
|
NM_004574
|
SEPT4
|
septin 4
|
|
chr20_-_33999760
|
4.447
|
|
UQCC
|
ubiquinol-cytochrome c reductase complex chaperone
|
|
chr2_+_210288668
|
4.402
|
NM_001039538
|
MAP2
|
microtubule-associated protein 2
|
|
chr5_-_42811959
|
4.336
|
NM_001085486
NM_001093726
NM_005410
|
SEPP1
|
selenoprotein P, plasma, 1
|
|
chr6_+_30594607
|
4.291
|
NM_024909
NM_001031722
NM_001190724
|
ATAT1
|
alpha tubulin acetyltransferase 1
|
|
chr9_-_138853146
|
4.231
|
NM_016172
|
UBAC1
|
UBA domain containing 1
|
|
chr2_-_110874142
|
4.181
|
NM_005434
|
MALL
|
mal, T-cell differentiation protein-like
|
|
chr7_-_143059144
|
4.164
|
NM_014690
|
FAM131B
|
family with sequence similarity 131, member B
|
|
chr10_+_123872440
|
4.135
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr12_-_57039785
|
4.124
|
NM_001686
|
ATP5B
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide
|
|
chr14_-_103989195
|
4.118
|
NM_001823
|
CKB
|
creatine kinase, brain
|
|
chr12_-_52867521
|
4.104
|
NM_173086
|
KRT6C
|
keratin 6C
|
|
chr19_-_55668956
|
4.081
|
NM_000363
|
TNNI3
|
troponin I type 3 (cardiac)
|
|
chr1_+_47603083
|
4.081
|
NM_001010969
|
CYP4A22
|
cytochrome P450, family 4, subfamily A, polypeptide 22
|
|
chr17_-_18218203
|
4.071
|
NM_004618
|
TOP3A
|
topoisomerase (DNA) III alpha
|
|
chr11_-_6462129
|
4.067
|
NM_000613
|
HPX
|
hemopexin
|
|
chr16_-_68269948
|
4.058
|
NM_024939
|
ESRP2
|
epithelial splicing regulatory protein 2
|
|
chr19_-_58067724
|
4.056
|
NM_001039654
|
ZNF550
|
zinc finger protein 550
|
|
chr20_-_48532036
|
4.040
|
|
SPATA2
|
spermatogenesis associated 2
|
|
chr2_-_152955810
|
4.038
|
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr3_+_129247481
|
4.029
|
NM_000539
|
RHO
|
rhodopsin
|
|
chr12_+_98987674
|
4.021
|
|
SLC25A3
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3
|
|
chr15_+_59908908
|
4.018
|
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type
|
|
chr1_+_54359859
|
3.993
|
NM_000792
NM_001039715
NM_001039716
NM_213593
|
DIO1
|
deiodinase, iodothyronine, type I
|
|
chr3_+_159557649
|
3.978
|
NM_001197109
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr2_-_110873638
|
3.943
|
|
MALL
|
mal, T-cell differentiation protein-like
|
|
chr17_-_2614926
|
3.926
|
NM_015229
|
KIAA0664
|
KIAA0664
|
|
chr11_-_66139064
|
3.906
|
NM_001532
|
SLC29A2
|
solute carrier family 29 (nucleoside transporters), member 2
|
|
chr3_+_52813936
|
3.900
|
NM_001166435
NM_001166436
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1
|
|
chr10_+_105036783
|
3.895
|
NM_032727
|
INA
|
internexin neuronal intermediate filament protein, alpha
|
|
chr18_-_74729049
|
3.892
|
NM_001025081
NM_001025090
NM_001025092
NM_002385
|
MBP
|
myelin basic protein
|
|
chr6_-_46293628
|
3.877
|
NM_005822
|
RCAN2
|
regulator of calcineurin 2
|
|
chr19_+_6739697
|
3.850
|
NM_004240
|
TRIP10
|
thyroid hormone receptor interactor 10
|
|
chr16_+_8807426
|
3.839
|
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr10_-_50970321
|
3.833
|
NM_001143996
NM_001143997
NM_018245
|
OGDHL
|
oxoglutarate dehydrogenase-like
|
|
chr22_+_32149936
|
3.830
|
NM_001242897
|
DEPDC5
|
DEP domain containing 5
|
|
chr16_+_69796237
|
3.825
|
NM_007014
NM_199423
|
WWP2
|
WW domain containing E3 ubiquitin protein ligase 2
|
|
chr19_-_51869632
|
3.821
|
NM_001985
|
ETFB
|
electron-transfer-flavoprotein, beta polypeptide
|
|
chr9_-_138853007
|
3.819
|
|
UBAC1
|
UBA domain containing 1
|
|
chr11_-_107582733
|
3.788
|
NM_003063
|
SLN
|
sarcolipin
|
|
chr5_+_161274196
|
3.788
|
NM_000806
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr1_-_230849863
|
3.783
|
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr16_+_28891234
|
3.782
|
|
ATP2A1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1
|
|
chr1_+_233749917
|
3.777
|
|
KCNK1
|
potassium channel, subfamily K, member 1
|
|
chr1_+_46640748
|
3.743
|
NM_005727
|
TSPAN1
|
tetraspanin 1
|
|
chr3_+_133464883
|
3.735
|
NM_001063
|
TF
|
transferrin
|
|
chr20_-_48531986
|
3.730
|
|
SPATA2
|
spermatogenesis associated 2
|
|
chr10_-_123353771
|
3.719
|
NM_001144916
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr3_+_132379130
|
3.710
|
NM_024818
|
UBA5
|
ubiquitin-like modifier activating enzyme 5
|
|
chr19_-_51472003
|
3.699
|
NM_001012964
NM_001012965
|
KLK6
|
kallikrein-related peptidase 6
|
|
chr22_-_29711650
|
3.695
|
NM_001007279
NM_006477
|
RASL10A
|
RAS-like, family 10, member A
|
|
chr1_-_157108169
|
3.693
|
NM_005240
NM_001145312
|
ETV3
|
ets variant 3
|
|
chr18_-_43678251
|
3.669
|
|
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle
|
|
chr19_-_15235748
|
3.665
|
|
ILVBL
|
ilvB (bacterial acetolactate synthase)-like
|
|
chr11_+_47270433
|
3.657
|
NM_001130102
|
NR1H3
|
nuclear receptor subfamily 1, group H, member 3
|
|
chr10_-_101841593
|
3.646
|
NM_001308
|
CPN1
|
carboxypeptidase N, polypeptide 1
|
|
chr11_+_67798077
|
3.640
|
NM_002496
|
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase)
|
|
chr2_-_152590983
|
3.636
|
NM_001164507
NM_001164508
NM_004543
|
NEB
|
nebulin
|
|
chr2_-_21225640
|
3.634
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr12_-_81991959
|
3.627
|
NM_001220477
NM_001220478
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr11_-_624901
|
3.620
|
NM_001171968
NM_021924
NM_031264
|
CDHR5
|
cadherin-related family member 5
|
|
chr12_-_122296619
|
3.616
|
NM_002150
|
HPD
|
4-hydroxyphenylpyruvate dioxygenase
|
|
chr2_-_110873353
|
3.600
|
|
MALL
|
mal, T-cell differentiation protein-like
|
|
chr22_-_29711474
|
3.595
|
|
RASL10A
|
RAS-like, family 10, member A
|
|
chr5_-_176831385
|
3.565
|
|
F12
|
coagulation factor XII (Hageman factor)
|
|
chr1_+_161172125
|
3.548
|
|
NDUFS2
|
NADH dehydrogenase (ubiquinone) Fe-S protein 2, 49kDa (NADH-coenzyme Q reductase)
|
|
chr15_+_78441737
|
3.540
|
|
IDH3A
|
isocitrate dehydrogenase 3 (NAD+) alpha
|
|
chr7_+_45614081
|
3.536
|
NM_021116
|
ADCY1
|
adenylate cyclase 1 (brain)
|
|
chr7_-_10979752
|
3.523
|
NM_002489
|
NDUFA4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4, 9kDa
|
|
chr18_-_43678195
|
3.502
|
|
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle
|
|
chr11_+_1860718
|
3.500
|
NM_001145829
|
TNNI2
|
troponin I type 2 (skeletal, fast)
|
|
chr18_-_43678230
|
3.484
|
|
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle
|
|
chr10_+_105037441
|
3.470
|
|
INA
|
internexin neuronal intermediate filament protein, alpha
|
|
chr12_-_57039736
|
3.467
|
|
ATP5B
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide
|
|
chr8_+_21916858
|
3.447
|
|
EPB49
|
erythrocyte membrane protein band 4.9 (dematin)
|
|
chr16_+_103827
|
3.423
|
NM_024571
|
SNRNP25
|
small nuclear ribonucleoprotein 25kDa (U11/U12)
|
|
chr12_-_56106061
|
3.420
|
NM_001144997
|
ITGA7
|
integrin, alpha 7
|
|
chr5_-_149792370
|
3.411
|
NM_001025158
NM_001025159
NM_004355
|
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain
|
|
chr18_-_43678233
|
3.401
|
|
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle
|
|
chr3_-_52312257
|
3.379
|
|
WDR82
|
WD repeat domain 82
|
|
chr16_+_103859
|
3.379
|
|
SNRNP25
|
small nuclear ribonucleoprotein 25kDa (U11/U12)
|
|
chr14_+_93389491
|
3.379
|
|
CHGA
|
chromogranin A (parathyroid secretory protein 1)
|
|
chr11_-_63933204
|
3.343
|
|
MACROD1
|
MACRO domain containing 1
|
|
chr1_+_20964414
|
3.342
|
|
|
|
|
chr16_-_89768094
|
3.324
|
NM_152339
|
SPATA2L
|
spermatogenesis associated 2-like
|
|
chr4_-_40517913
|
3.323
|
NM_019027
|
RBM47
|
RNA binding motif protein 47
|
|
chr7_+_86273219
|
3.313
|
NM_000840
|
GRM3
|
glutamate receptor, metabotropic 3
|
|
chr10_-_75634219
|
3.303
|
NM_001204492
NM_001222
NM_172169
NM_172170
NM_172171
NM_172173
|
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma
|
|
chr19_-_51568323
|
3.303
|
NM_015596
|
KLK13
|
kallikrein-related peptidase 13
|
|
chr1_+_44412582
|
3.299
|
|
IPO13
|
importin 13
|
|
chr19_-_36246411
|
3.274
|
|
HSPB6
|
heat shock protein, alpha-crystallin-related, B6
|
|
chr1_-_201346804
|
3.267
|
NM_000364
NM_001001430
NM_001001431
NM_001001432
|
TNNT2
|
troponin T type 2 (cardiac)
|
|
chr11_-_111782447
|
3.265
|
NM_001885
|
CRYAB
|
crystallin, alpha B
|
|
chr17_-_7166263
|
3.247
|
NM_001185023
NM_001307
|
CLDN7
|
claudin 7
|
|
chr3_+_39509069
|
3.246
|
NM_182935
|
MOBP
|
myelin-associated oligodendrocyte basic protein
|
|
chr11_+_17757494
|
3.239
|
NM_001112741
NM_004976
|
KCNC1
|
potassium voltage-gated channel, Shaw-related subfamily, member 1
|
|
chr11_+_117947726
|
3.225
|
NM_001083947
NM_001173551
NM_001173552
NM_019894
|
TMPRSS4
|
transmembrane protease, serine 4
|
|
chr19_-_15235967
|
3.215
|
|
ILVBL
|
ilvB (bacterial acetolactate synthase)-like
|
|
chr4_+_76439653
|
3.213
|
NM_144721
|
THAP6
|
THAP domain containing 6
|
|
chrX_+_135230736
|
3.205
|
NM_001159700
|
FHL1
|
four and a half LIM domains 1
|
|
chr4_+_159593233
|
3.204
|
NM_004453
|
ETFDH
|
electron-transferring-flavoprotein dehydrogenase
|
|
chr1_-_27701302
|
3.195
|
NM_003665
NM_173452
|
FCN3
|
ficolin (collagen/fibrinogen domain containing) 3 (Hakata antigen)
|
|
chr18_-_43678257
|
3.175
|
|
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle
|