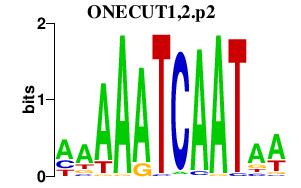
|
chr5_-_149792370
|
23.356
|
NM_001025158
NM_001025159
NM_004355
|
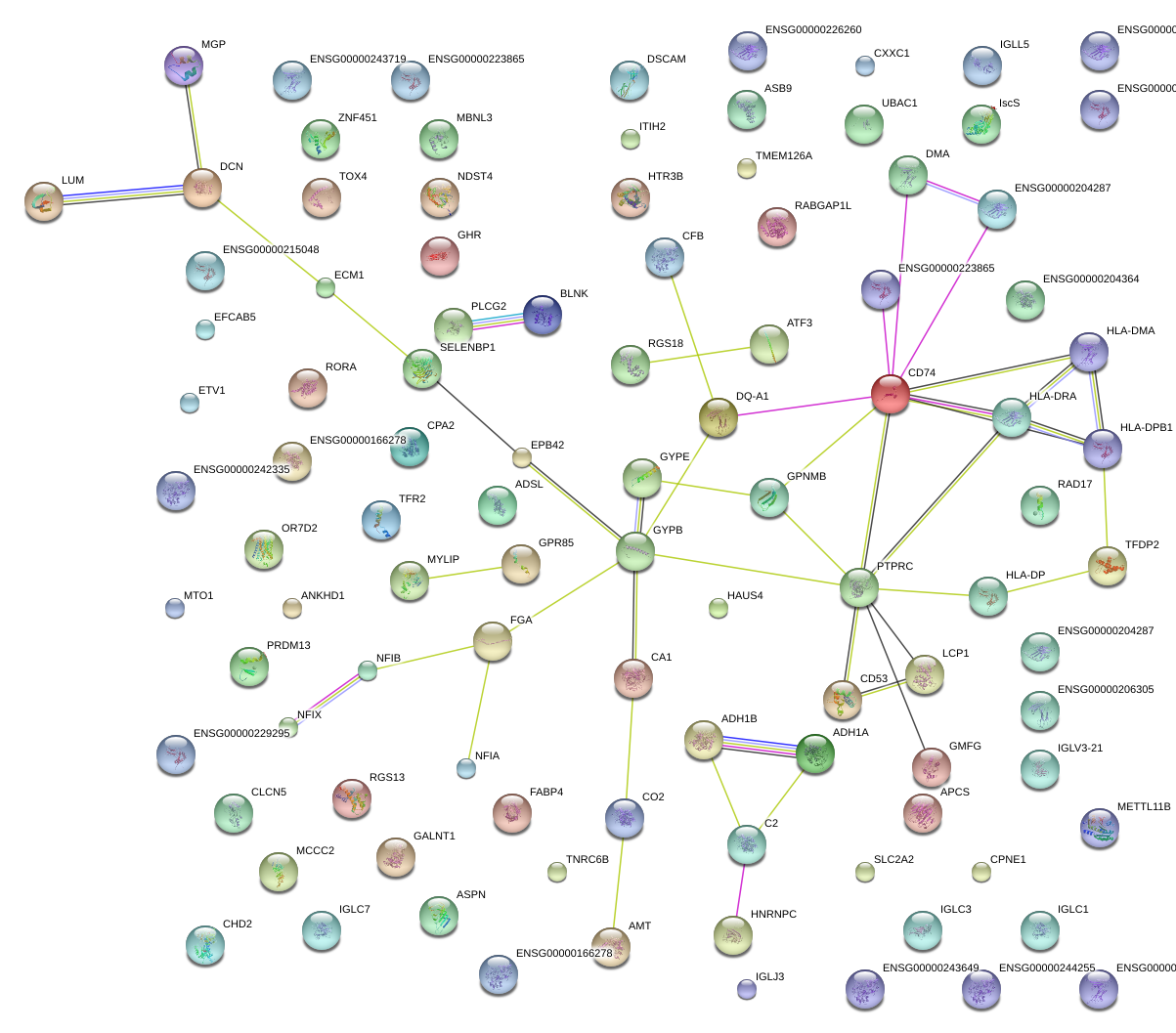
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain
|
|
chr5_-_149792276
|
22.253
|
|
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain
|
|
chr22_+_23054862
|
21.123
|
|
IGLV3-21
IGLJ3
IGLC1
|
immunoglobulin lambda variable 3-21
immunoglobulin lambda joining 3
immunoglobulin lambda constant 1 (Mcg marker)
|
|
chr2_-_89165652
|
19.592
|
|
|
|
|
chr6_+_33043702
|
17.127
|
NM_002121
|
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1
|
|
chr12_-_91573264
|
12.204
|
NM_133503
|
DCN
|
decorin
|
|
chr1_+_198608136
|
10.708
|
NM_002838
NM_080921
NM_080923
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr7_+_23286420
|
10.501
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr7_+_23286384
|
10.258
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr1_-_151345156
|
10.167
|
NM_003944
|
SELENBP1
|
selenium binding protein 1
|
|
chr10_-_98031154
|
10.006
|
NM_001114094
NM_013314
|
BLNK
|
B-cell linker
|
|
chr4_-_100212085
|
9.978
|
NM_000667
|
ADH1A
|
alcohol dehydrogenase 1A (class I), alpha polypeptide
|
|
chr1_+_198608244
|
9.940
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr18_-_47814691
|
9.733
|
NM_001101654
NM_014593
|
CXXC1
|
CXXC finger protein 1
|
|
chr15_-_43513024
|
9.701
|
|
EPB42
|
erythrocyte membrane protein band 4.2
|
|
chr1_+_198608216
|
8.660
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr15_-_43513234
|
8.573
|
NM_000119
NM_001114134
|
EPB42
|
erythrocyte membrane protein band 4.2
|
|
chr7_-_14025826
|
8.369
|
NM_001163150
NM_001163151
NM_001163152
|
ETV1
|
ets variant 1
|
|
chr8_-_82395401
|
8.331
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr3_-_141747434
|
8.277
|
NM_001178141
NM_001178142
NM_006286
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr15_-_43513196
|
8.047
|
|
EPB42
|
erythrocyte membrane protein band 4.2
|
|
chr7_+_23286386
|
8.004
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr6_-_32920812
|
7.996
|
NM_006120
|
HLA-DMA
|
major histocompatibility complex, class II, DM alpha
|
|
chr12_-_15038724
|
7.736
|
NM_000900
NM_001190839
|
MGP
|
matrix Gla protein
|
|
chr1_+_111413820
|
7.562
|
NM_001040033
|
CD53
|
CD53 molecule
|
|
chr12_-_91576579
|
7.127
|
|
DCN
|
decorin
|
|
chr7_+_23286277
|
6.981
|
NM_001005340
NM_002510
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr14_-_23426356
|
6.768
|
|
HAUS4
|
HAUS augmin-like complex, subunit 4
|
|
chr1_+_192605267
|
6.732
|
NM_002927
NM_144766
|
RGS13
|
regulator of G-protein signaling 13
|
|
chr15_-_60919642
|
6.552
|
NM_002943
NM_134260
|
RORA
|
RAR-related orphan receptor A
|
|
chr6_+_31913852
|
6.389
|
|
CFB
|
complement factor B
|
|
chr8_-_86290333
|
6.077
|
NM_001128829
NM_001128830
NM_001128831
NM_001738
|
CA1
|
carbonic anhydrase I
|
|
chr4_-_155511838
|
5.853
|
NM_000508
NM_021871
|
FGA
|
fibrinogen alpha chain
|
|
chr4_-_144940439
|
5.815
|
NM_002100
|
GYPB
|
glycophorin B (MNS blood group)
|
|
chr13_-_46756295
|
5.753
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr12_-_91505215
|
5.729
|
|
LUM
|
lumican
|
|
chr7_+_23286390
|
5.533
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr16_+_81812897
|
5.487
|
NM_002661
|
PLCG2
|
phospholipase C, gamma 2 (phosphatidylinositol-specific)
|
|
chr7_-_100239136
|
5.470
|
NM_003227
|
TFR2
|
transferrin receptor 2
|
|
chr9_-_95244634
|
5.306
|
|
ASPN
|
asporin
|
|
chr6_+_32407618
|
5.298
|
NM_019111
|
HLA-DRA
|
major histocompatibility complex, class II, DR alpha
|
|
chr7_-_112726143
|
5.259
|
NM_001146266
NM_001146267
|
GPR85
|
G protein-coupled receptor 85
|
|
chr11_+_17332454
|
5.208
|
|
|
|
|
chr7_+_129906702
|
5.035
|
NM_001869
|
CPA2
|
carboxypeptidase A2 (pancreatic)
|
|
chrX_-_131573638
|
5.030
|
NM_018388
NM_133486
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr17_+_28268622
|
4.946
|
NM_198529
|
EFCAB5
|
EF-hand calcium binding domain 5
|
|
chr9_-_95244750
|
4.925
|
NM_001193335
NM_017680
|
ASPN
|
asporin
|
|
chr19_-_39826676
|
4.905
|
NM_004877
|
GMFG
|
glia maturation factor, gamma
|
|
chr3_-_49459870
|
4.903
|
|
AMT
|
aminomethyltransferase
|
|
chr5_+_139876504
|
4.830
|
|
ANKHD1
|
ankyrin repeat and KH domain containing 1
|
|
chr15_+_93443418
|
4.780
|
NM_001042572
NM_001271
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr16_+_81812899
|
4.755
|
|
PLCG2
|
phospholipase C, gamma 2 (phosphatidylinositol-specific)
|
|
chr14_-_23426265
|
4.605
|
NM_001166269
NM_001166270
NM_017815
|
HAUS4
|
HAUS augmin-like complex, subunit 4
|
|
chr10_+_7745341
|
4.551
|
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2
|
|
chr10_+_7745354
|
4.537
|
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2
|
|
chr3_-_49460110
|
4.533
|
NM_000481
NM_001164710
NM_001164711
NM_001164712
|
AMT
|
aminomethyltransferase
|
|
chr6_+_30227335
|
4.497
|
|
HLA-L
|
major histocompatibility complex, class I, L, pseudogene
|
|
chrX_-_15288172
|
4.398
|
NM_001031739
NM_001168530
NM_024087
|
ASB9
|
ankyrin repeat and SOCS box containing 9
|
|
chr12_-_91576805
|
4.395
|
NM_001920
|
DCN
|
decorin
|
|
chr5_+_115898218
|
4.375
|
|
|
|
|
chr10_+_7745229
|
4.346
|
NM_002216
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2
|
|
chr6_+_31895201
|
4.321
|
NM_000063
NM_001145903
|
C2
|
complement component 2
|
|
chr6_+_16143281
|
4.320
|
|
MYLIP
|
myosin regulatory light chain interacting protein
|
|
chr5_+_68667280
|
4.252
|
NM_133339
|
RAD17
|
RAD17 homolog (S. pombe)
|
|
chr10_+_7745325
|
4.206
|
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2
|
|
chrX_+_49832214
|
4.152
|
NM_000084
|
CLCN5
|
chloride channel 5
|
|
chr10_+_7745284
|
4.095
|
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2
|
|
chr4_-_116034932
|
3.941
|
NM_022569
|
NDST4
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4
|
|
chr6_+_74171610
|
3.931
|
|
MTO1
|
mitochondrial translation optimization 1 homolog (S. cerevisiae)
|
|
chr6_+_56954985
|
3.873
|
|
ZNF451
|
zinc finger protein 451
|
|
chr6_+_56954978
|
3.858
|
|
ZNF451
|
zinc finger protein 451
|
|
chr11_+_113775517
|
3.813
|
NM_006028
|
HTR3B
|
5-hydroxytryptamine (serotonin) receptor 3B
|
|
chr19_+_9296269
|
3.762
|
NM_175883
|
OR7D2
|
olfactory receptor, family 7, subfamily D, member 2
|
|
chr13_-_46756326
|
3.753
|
NM_002298
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr22_+_40573879
|
3.741
|
NM_001162501
NM_015088
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr9_-_138853146
|
3.707
|
NM_016172
|
UBAC1
|
UBA domain containing 1
|
|
chr3_-_170744767
|
3.685
|
NM_000340
|
SLC2A2
|
solute carrier family 2 (facilitated glucose transporter), member 2
|
|
chr6_+_100054649
|
3.662
|
NM_021620
|
PRDM13
|
PR domain containing 13
|
|
chr1_+_159557615
|
3.659
|
NM_001639
|
APCS
|
amyloid P component, serum
|
|
chr1_+_150480486
|
3.642
|
NM_001202858
NM_004425
NM_022664
|
ECM1
|
extracellular matrix protein 1
|
|
chr6_+_56954889
|
3.618
|
|
ZNF451
|
zinc finger protein 451
|
|
chr9_-_14086901
|
3.615
|
|
NFIB
|
nuclear factor I/B
|
|
chr1_+_212788359
|
3.596
|
NM_001206486
|
ATF3
|
activating transcription factor 3
|
|
chr5_+_70944968
|
3.562
|
|
MCCC2
|
methylcrotonoyl-CoA carboxylase 2 (beta)
|
|
chr18_+_33234532
|
3.531
|
NM_020474
|
GALNT1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 (GalNAc-T1)
|
|
chr20_-_34241830
|
3.523
|
NM_003915
|
CPNE1
|
copine I
|
|
chr5_+_42548144
|
3.510
|
NM_001242402
|
GHR
|
growth hormone receptor
|
|
chr1_+_174843523
|
3.468
|
NM_001243763
|
RABGAP1L
|
RAB GTPase activating protein 1-like
|
|
chr10_-_116392099
|
3.455
|
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr11_-_5255712
|
3.439
|
NM_000519
|
HBD
|
hemoglobin, delta
|
|
chr7_+_80267922
|
3.439
|
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chrX_+_46940235
|
3.324
|
|
RGN
|
regucalcin (senescence marker protein-30)
|
|
chr10_+_89267589
|
3.288
|
NM_001178118
|
MINPP1
|
multiple inositol-polyphosphate phosphatase 1
|
|
chr1_+_63063186
|
3.265
|
NM_014495
|
ANGPTL3
|
angiopoietin-like 3
|
|
chr12_+_54892567
|
3.257
|
NM_001184976
|
NCKAP1L
|
NCK-associated protein 1-like
|
|
chr5_-_98262217
|
3.247
|
NM_001270
|
CHD1
|
chromodomain helicase DNA binding protein 1
|
|
chr10_+_124320180
|
3.241
|
NM_004406
NM_007329
NM_017579
|
DMBT1
|
deleted in malignant brain tumors 1
|
|
chr18_+_72922725
|
3.198
|
NM_005786
|
TSHZ1
|
teashirt zinc finger homeobox 1
|
|
chr9_-_138853007
|
3.194
|
|
UBAC1
|
UBA domain containing 1
|
|
chr2_+_172821874
|
3.181
|
|
HAT1
|
histone acetyltransferase 1
|
|
chr6_+_56954801
|
3.146
|
NM_001031623
NM_015555
|
ZNF451
|
zinc finger protein 451
|
|
chr8_-_108510094
|
3.124
|
NM_001146
NM_001199859
|
ANGPT1
|
angiopoietin 1
|
|
chrX_-_15288588
|
3.114
|
NM_001168531
|
ASB9
|
ankyrin repeat and SOCS box containing 9
|
|
chr3_+_100238063
|
3.074
|
|
TMEM45A
|
transmembrane protein 45A
|
|
chr6_+_34204928
|
3.021
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr1_+_15764934
|
2.990
|
NM_007272
|
CTRC
|
chymotrypsin C (caldecrin)
|
|
chr1_-_104238785
|
2.961
|
NM_004038
NM_001008218
NM_001008219
|
AMY1A
AMY1B
AMY1C
|
amylase, alpha 1A (salivary)
amylase, alpha 1B (salivary)
amylase, alpha 1C (salivary)
|
|
chr13_-_46756291
|
2.961
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr4_-_151770611
|
2.958
|
|
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing
|
|
chr5_+_68666861
|
2.957
|
NM_133340
NM_133341
|
RAD17
|
RAD17 homolog (S. pombe)
|
|
chr5_-_160279045
|
2.938
|
NM_025153
|
ATP10B
|
ATPase, class V, type 10B
|
|
chr14_+_21457964
|
2.933
|
NM_001029991
NM_022734
|
METTL17
|
methyltransferase like 17
|
|
chr12_+_32654976
|
2.925
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr6_+_30130982
|
2.885
|
NM_033229
|
TRIM15
|
tripartite motif containing 15
|
|
chr8_-_93111915
|
2.867
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr2_-_79386878
|
2.809
|
NM_138938
NM_002580
NM_138937
|
REG3A
|
regenerating islet-derived 3 alpha
|
|
chr22_-_37098620
|
2.794
|
NM_006078
|
CACNG2
|
calcium channel, voltage-dependent, gamma subunit 2
|
|
chr1_+_212209129
|
2.767
|
|
DTL
|
denticleless homolog (Drosophila)
|
|
chr1_-_158656487
|
2.759
|
NM_003126
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2)
|
|
chr12_-_4488707
|
2.728
|
NM_020638
|
FGF23
|
fibroblast growth factor 23
|
|
chr12_+_41831484
|
2.704
|
NM_013377
|
PDZRN4
|
PDZ domain containing ring finger 4
|
|
chr17_-_49124128
|
2.675
|
NM_001251971
|
SPAG9
|
sperm associated antigen 9
|
|
chr1_+_104198140
|
2.652
|
NM_001008221
|
AMY1A
|
amylase, alpha 1A (salivary)
|
|
chr6_-_64029847
|
2.639
|
NM_001143940
NM_016571
|
LGSN
|
lengsin, lens protein with glutamine synthetase domain
|
|
chr1_-_217262946
|
2.629
|
NM_001243506
NM_206594
NM_206595
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr1_+_104292439
|
2.619
|
NM_004038
NM_001008218
NM_001008219
|
AMY1A
AMY1B
AMY1C
|
amylase, alpha 1A (salivary)
amylase, alpha 1B (salivary)
amylase, alpha 1C (salivary)
|
|
chr1_+_104292278
|
2.606
|
NM_001008221
|
AMY1A
|
amylase, alpha 1A (salivary)
|
|
chr16_-_15833937
|
2.590
|
|
MYH11
|
myosin, heavy chain 11, smooth muscle
|
|
chr11_+_22696318
|
2.580
|
NM_005256
|
GAS2
|
growth arrest-specific 2
|
|
chr12_-_102512277
|
2.567
|
NM_024057
|
NUP37
|
nucleoporin 37kDa
|
|
chr4_-_69536314
|
2.556
|
NM_001076
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15
|
|
chr21_-_43735705
|
2.544
|
NM_003226
|
TFF3
|
trefoil factor 3 (intestinal)
|
|
chr1_-_104239071
|
2.529
|
NM_001008221
|
AMY1A
|
amylase, alpha 1A (salivary)
|
|
chr1_+_24018293
|
2.528
|
|
RPL11
|
ribosomal protein L11
|
|
chr13_-_46679143
|
2.486
|
NM_001872
NM_016413
|
CPB2
|
carboxypeptidase B2 (plasma)
|
|
chr14_+_37126772
|
2.478
|
NM_006194
|
PAX9
|
paired box 9
|
|
chr6_+_45296053
|
2.478
|
NM_001015051
NM_001024630
|
RUNX2
|
runt-related transcription factor 2
|
|
chr3_-_105421044
|
2.464
|
|
CBLB
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence b
|
|
chr1_+_104198301
|
2.462
|
NM_004038
NM_001008218
NM_001008219
|
AMY1A
AMY1B
AMY1C
|
amylase, alpha 1A (salivary)
amylase, alpha 1B (salivary)
amylase, alpha 1C (salivary)
|
|
chr11_+_57529233
|
2.423
|
NM_001085458
NM_001085459
NM_001085460
NM_001085461
NM_001085462
NM_001085463
NM_001085464
NM_001085465
NM_001085466
NM_001085467
NM_001085468
NM_001085469
NM_001206883
NM_001206884
NM_001206885
NM_001206886
NM_001206887
NM_001206888
NM_001206889
NM_001206890
NM_001206891
NM_001331
|
CTNND1
|
catenin (cadherin-associated protein), delta 1
|
|
chr16_+_81812925
|
2.406
|
|
PLCG2
|
phospholipase C, gamma 2 (phosphatidylinositol-specific)
|
|
chr13_-_103719195
|
2.397
|
NM_000452
|
SLC10A2
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 2
|
|
chr17_-_3599298
|
2.390
|
|
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5
|
|
chr7_+_80267782
|
2.382
|
NM_000072
NM_001001548
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr3_+_152017193
|
2.310
|
NM_207293
NM_207294
NM_207295
NM_207296
NM_207297
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr10_-_124605690
|
2.304
|
NM_022034
|
CUZD1
|
CUB and zona pellucida-like domains 1
|
|
chr16_-_18798077
|
2.288
|
|
|
|
|
chr7_+_45933096
|
2.286
|
|
|
|
|
chr14_-_24020766
|
2.277
|
NM_033400
|
ZFHX2
|
zinc finger homeobox 2
|
|
chr9_-_123812535
|
2.268
|
NM_001735
|
C5
|
complement component 5
|
|
chr19_-_14992166
|
2.266
|
NM_030901
|
OR7A17
|
olfactory receptor, family 7, subfamily A, member 17
|
|
chr17_-_38721701
|
2.218
|
NM_001838
|
CCR7
|
chemokine (C-C motif) receptor 7
|
|
chr4_+_8951476
|
2.190
|
NM_001040071
|
LOC650293
|
seven transmembrane helix receptor
|
|
chr12_+_21525801
|
2.185
|
NM_000415
|
IAPP
|
islet amyloid polypeptide
|
|
chr17_-_3599359
|
2.167
|
|
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5
|
|
chr22_+_22723972
|
2.165
|
|
|
|
|
chr12_-_91398757
|
2.163
|
NM_004950
|
EPYC
|
epiphycan
|
|
chr10_-_65028825
|
2.144
|
NM_004241
|
JMJD1C
|
jumonji domain containing 1C
|
|
chr15_-_80263460
|
2.144
|
NM_001114735
NM_004049
|
BCL2A1
|
BCL2-related protein A1
|
|
chr4_-_70080448
|
2.128
|
NM_001073
|
UGT2B11
UGT2B10
|
UDP glucuronosyltransferase 2 family, polypeptide B11
UDP glucuronosyltransferase 2 family, polypeptide B10
|
|
chr4_-_84035888
|
2.101
|
|
PLAC8
|
placenta-specific 8
|
|
chr15_+_74900712
|
2.099
|
NM_003992
|
CLK3
|
CDC-like kinase 3
|
|
chr3_+_148545587
|
2.092
|
NM_001871
|
CPB1
|
carboxypeptidase B1 (tissue)
|
|
chr2_+_79252825
|
2.091
|
NM_001008387
NM_198448
|
REG3G
|
regenerating islet-derived 3 gamma
|
|
chr2_+_143886969
|
2.079
|
|
ARHGAP15
|
Rho GTPase activating protein 15
|
|
chr19_-_5846465
|
2.070
|
|
FUT3
|
fucosyltransferase 3 (galactoside 3(4)-L-fucosyltransferase, Lewis blood group)
|
|
chrX_+_100474932
|
2.064
|
NM_001171184
NM_001939
|
DRP2
|
dystrophin related protein 2
|
|
chrX_-_64196268
|
2.041
|
NM_001178033
NM_018684
|
ZC4H2
|
zinc finger, C4H2 domain containing
|
|
chr6_-_79675672
|
2.027
|
|
PHIP
|
pleckstrin homology domain interacting protein
|
|
chr1_+_45477916
|
2.026
|
|
UROD
|
uroporphyrinogen decarboxylase
|
|
chr3_-_57234279
|
2.020
|
NM_003865
|
HESX1
|
HESX homeobox 1
|
|
chr1_-_116383323
|
2.002
|
NM_001111061
|
NHLH2
|
nescient helix loop helix 2
|
|
chr4_-_69886112
|
1.995
|
NM_001075
NM_001144767
|
UGT2B10
|
UDP glucuronosyltransferase 2 family, polypeptide B10
|
|
chr4_-_72649734
|
1.995
|
NM_000583
|
GC
|
group-specific component (vitamin D binding protein)
|
|
chr6_-_32634287
|
1.994
|
NM_001243961
NM_002123
|
HLA-DQB1
|
major histocompatibility complex, class II, DQ beta 1
|
|
chr1_+_117297056
|
1.991
|
NM_001767
|
CD2
|
CD2 molecule
|
|
chr10_+_71561687
|
1.989
|
|
COL13A1
|
collagen, type XIII, alpha 1
|
|
chr12_-_9268440
|
1.986
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr1_+_24018268
|
1.985
|
NM_000975
NM_001199802
|
RPL11
|
ribosomal protein L11
|
|
chr16_+_69680959
|
1.981
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr14_+_58894709
|
1.956
|
NM_014749
|
KIAA0586
|
KIAA0586
|
|
chr4_+_69681709
|
1.934
|
NM_001075
NM_001144767
|
UGT2B10
|
UDP glucuronosyltransferase 2 family, polypeptide B10
|
|
chr3_+_186383770
|
1.928
|
NM_000412
|
HRG
|
histidine-rich glycoprotein
|
|
chr11_-_128712340
|
1.926
|
NM_000220
|
KCNJ1
|
potassium inwardly-rectifying channel, subfamily J, member 1
|
|
chrX_-_24690740
|
1.909
|
NM_001163264
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr11_+_94800055
|
1.864
|
NM_032102
|
SRSF8
|
serine/arginine-rich splicing factor 8
|
|
chr8_+_17396285
|
1.857
|
NM_001164771
NM_003046
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr17_-_3337134
|
1.853
|
NM_003554
|
OR1E2
|
olfactory receptor, family 1, subfamily E, member 2
|
|
chr6_+_27925018
|
1.850
|
NM_012367
|
OR2B6
|
olfactory receptor, family 2, subfamily B, member 6
|
|
chr2_-_217540722
|
1.829
|
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr8_+_123794043
|
1.816
|
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr4_+_70916158
|
1.815
|
NM_002159
|
HTN1
|
histatin 1
|
|
chr7_+_80267972
|
1.813
|
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr10_-_29754478
|
1.811
|
|
SVIL
|
supervillin
|
|
chr8_-_87755880
|
1.804
|
NM_019098
|
CNGB3
|
cyclic nucleotide gated channel beta 3
|
|
chr20_+_32150139
|
1.799
|
NM_001039709
NM_005093
|
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2
|
|
chr2_-_179672056
|
1.795
|
NM_003319
NM_133378
NM_133379
NM_133432
NM_133437
|
TTN
|
titin
|
|
chr15_-_64673569
|
1.795
|
NM_001029989
NM_014736
|
KIAA0101
|
KIAA0101
|
|
chr7_+_129008044
|
1.794
|
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr7_-_25268057
|
1.763
|
NM_022150
|
NPVF
|
neuropeptide VF precursor
|