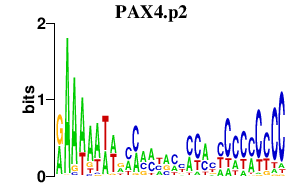
|
chr4_-_176733413
|
58.417
|
|
GPM6A
|
glycoprotein M6A
|
|
chr4_-_176733901
|
36.478
|
|
GPM6A
|
glycoprotein M6A
|
|
chr1_+_151032150
|
25.694
|
NM_006818
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr1_+_151032880
|
24.456
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chrX_+_135067634
|
24.379
|
|
SLC9A6
|
solute carrier family 9 (sodium/hydrogen exchanger), member 6
|
|
chr1_+_151032683
|
24.185
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chrX_+_135067573
|
20.808
|
NM_001177651
NM_001042537
NM_006359
|
SLC9A6
|
solute carrier family 9 (sodium/hydrogen exchanger), member 6
|
|
chrX_+_103031753
|
20.668
|
NM_000533
NM_199478
|
PLP1
|
proteolipid protein 1
|
|
chr6_+_32407618
|
19.718
|
NM_019111
|
HLA-DRA
|
major histocompatibility complex, class II, DR alpha
|
|
chr8_+_41348105
|
19.687
|
|
GOLGA7
|
golgin A7
|
|
chr8_+_41348126
|
18.989
|
NM_001002296
|
GOLGA7
|
golgin A7
|
|
chr15_+_66694216
|
18.753
|
|
MAP2K1
|
mitogen-activated protein kinase kinase 1
|
|
chr8_+_41348094
|
18.442
|
|
GOLGA7
|
golgin A7
|
|
chrX_+_103031433
|
17.836
|
NM_001128834
|
PLP1
|
proteolipid protein 1
|
|
chr12_-_16759531
|
16.709
|
NM_018640
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr17_-_8063946
|
16.339
|
|
VAMP2
|
vesicle-associated membrane protein 2 (synaptobrevin 2)
|
|
chr1_-_1623942
|
16.203
|
|
SLC35E2B
SLC35E2
|
solute carrier family 35, member E2B
solute carrier family 35, member E2
|
|
chr11_-_62474666
|
15.319
|
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chr16_+_15528324
|
14.607
|
NM_033201
|
C16orf45
|
chromosome 16 open reading frame 45
|
|
chr11_-_62474869
|
14.393
|
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chr11_-_62474810
|
14.323
|
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chr8_+_41348067
|
14.312
|
NM_001174124
NM_016099
|
GOLGA7
|
golgin A7
|
|
chr11_-_62472901
|
14.300
|
|
|
|
|
chr7_+_69064316
|
14.031
|
|
AUTS2
|
autism susceptibility candidate 2
|
|
chr9_+_137967366
|
13.835
|
|
OLFM1
|
olfactomedin 1
|
|
chr5_+_36608430
|
13.798
|
NM_001166695
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr22_+_23229959
|
13.456
|
NM_001178126
|
IGLL5
IGLC1
|
immunoglobulin lambda-like polypeptide 5
immunoglobulin lambda constant 1 (Mcg marker)
|
|
chr7_+_80267972
|
13.423
|
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr5_-_146460991
|
13.412
|
NM_181677
NM_181678
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta
|
|
chr1_-_160681552
|
12.726
|
NM_001778
|
CD48
|
CD48 molecule
|
|
chr5_-_142783059
|
12.442
|
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr2_-_136873740
|
12.353
|
NM_001008540
|
CXCR4
|
chemokine (C-X-C motif) receptor 4
|
|
chr6_+_31582978
|
12.048
|
NM_004847
NM_001623
|
AIF1
|
allograft inflammatory factor 1
|
|
chr1_-_149889362
|
12.028
|
NM_014849
|
SV2A
|
synaptic vesicle glycoprotein 2A
|
|
chr6_+_31553955
|
11.819
|
NM_205838
NM_205839
|
LST1
|
leukocyte specific transcript 1
|
|
chr1_-_182357871
|
11.678
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr18_-_53089722
|
11.310
|
NM_001243233
|
TCF4
|
transcription factor 4
|
|
chr12_-_16760935
|
11.232
|
NM_001001395
NM_001243609
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr1_-_182357757
|
11.125
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr18_-_53255734
|
11.037
|
NM_001083962
NM_001243230
NM_003199
|
TCF4
|
transcription factor 4
|
|
chr19_-_54784922
|
11.033
|
NM_001080978
NM_005874
|
LILRB2
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 2
|
|
chr1_-_182360889
|
10.881
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr1_-_1624180
|
10.313
|
NM_001110781
|
SLC35E2B
|
solute carrier family 35, member E2B
|
|
chr9_+_19049424
|
10.286
|
|
RRAGA
|
Ras-related GTP binding A
|
|
chr9_+_19049408
|
10.285
|
|
RRAGA
|
Ras-related GTP binding A
|
|
chr7_+_26331611
|
10.154
|
|
SNX10
|
sorting nexin 10
|
|
chr8_+_86376130
|
10.097
|
NM_000067
|
CA2
|
carbonic anhydrase II
|
|
chr3_+_69134057
|
10.023
|
NM_006407
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5
|
|
chr6_+_31554475
|
10.021
|
NM_205837
|
LST1
|
leukocyte specific transcript 1
|
|
chr9_+_19049417
|
9.925
|
|
RRAGA
|
Ras-related GTP binding A
|
|
chr1_-_153348057
|
9.923
|
NM_005621
|
S100A12
|
S100 calcium binding protein A12
|
|
chr9_+_19049376
|
9.811
|
|
RRAGA
|
Ras-related GTP binding A
|
|
chr7_+_80267922
|
9.763
|
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr17_+_16284350
|
9.644
|
NM_018955
|
UBB
|
ubiquitin B
|
|
chr4_+_113739203
|
9.548
|
NM_001127493
|
ANK2
|
ankyrin 2, neuronal
|
|
chr3_+_238274
|
9.315
|
NM_001253387
NM_006614
|
CHL1
|
cell adhesion molecule with homology to L1CAM (close homolog of L1)
|
|
chr19_+_55141907
|
9.098
|
NM_001081638
NM_001081639
NM_001081637
|
LILRB1
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 1
|
|
chr1_+_101702304
|
9.029
|
NM_001400
|
S1PR1
|
sphingosine-1-phosphate receptor 1
|
|
chr9_+_19049355
|
8.997
|
NM_006570
|
RRAGA
|
Ras-related GTP binding A
|
|
chr12_+_32112381
|
8.844
|
|
C12orf35
|
chromosome 12 open reading frame 35
|
|
chr1_-_156399183
|
8.832
|
NM_006365
|
C1orf61
|
chromosome 1 open reading frame 61
|
|
chr1_+_101702561
|
8.658
|
|
S1PR1
|
sphingosine-1-phosphate receptor 1
|
|
chr11_-_66115016
|
8.467
|
NM_006876
|
B3GNT1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1
|
|
chr1_+_110543548
|
8.399
|
NM_001242674
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr9_-_91793681
|
8.367
|
NM_016848
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr4_-_84030995
|
8.335
|
NM_001130716
|
PLAC8
|
placenta-specific 8
|
|
chr12_-_16759430
|
8.323
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chrX_+_47004663
|
7.891
|
|
RBM10
|
RNA binding motif protein 10
|
|
chr1_-_182360181
|
7.726
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr6_+_32605182
|
7.719
|
NM_002122
|
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1
|
|
chrX_+_110187468
|
7.709
|
NM_001128166
NM_001128167
|
PAK3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr20_-_4795768
|
7.709
|
NM_170774
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr9_+_137967088
|
7.591
|
NM_014279
NM_006334
|
OLFM1
|
olfactomedin 1
|
|
chr16_-_28621279
|
7.543
|
NM_001055
NM_177529
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1
|
|
chr8_+_26435920
|
7.511
|
NM_001244604
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr4_+_41258941
|
7.492
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr3_+_238535
|
7.469
|
|
CHL1
|
cell adhesion molecule with homology to L1CAM (close homolog of L1)
|
|
chrX_+_47004633
|
7.455
|
|
RBM10
|
RNA binding motif protein 10
|
|
chr7_-_37488894
|
7.430
|
NM_001206480
|
ELMO1
|
engulfment and cell motility 1
|
|
chr7_-_124405029
|
7.399
|
|
GPR37
|
G protein-coupled receptor 37 (endothelin receptor type B-like)
|
|
chrX_+_47004670
|
7.363
|
|
RBM10
|
RNA binding motif protein 10
|
|
chr11_-_78052925
|
7.342
|
NM_012296
|
GAB2
|
GRB2-associated binding protein 2
|
|
chrX_+_47004665
|
7.298
|
|
RBM10
|
RNA binding motif protein 10
|
|
chrX_+_47004657
|
7.279
|
|
RBM10
|
RNA binding motif protein 10
|
|
chr12_-_16758839
|
7.236
|
NM_001243612
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chrX_+_47004676
|
7.200
|
|
RBM10
|
RNA binding motif protein 10
|
|
chr14_+_55738871
|
7.131
|
NM_152231
|
FBXO34
|
F-box protein 34
|
|
chr8_+_26435466
|
6.998
|
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr11_+_1874199
|
6.744
|
NM_002339
|
LSP1
|
lymphocyte-specific protein 1
|
|
chr8_-_21771111
|
6.629
|
NM_003974
|
DOK2
|
docking protein 2, 56kDa
|
|
chr6_-_32557431
|
6.560
|
NM_001243965
NM_002124
|
HLA-DRB1
HLA-DRB4
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 1
major histocompatibility complex, class II, DR beta 4
major histocompatibility complex, class II, DR beta 5
|
|
chr8_-_101571870
|
6.544
|
NM_198401
|
GAPDHP62
ANKRD46
|
glyceraldehyde 3 phosphate dehydrogenase pseudogene 62
ankyrin repeat domain 46
|
|
chr19_+_2977480
|
6.471
|
NM_001143986
NM_024760
|
TLE6
|
transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila)
|
|
chr4_+_41258909
|
6.292
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chrX_+_47004609
|
6.284
|
NM_001204466
NM_001204467
NM_001204468
NM_005676
NM_152856
|
RBM10
|
RNA binding motif protein 10
|
|
chr10_+_111967302
|
6.225
|
NM_130439
|
MXI1
|
MAX interactor 1
|
|
chr19_+_50015535
|
6.024
|
NM_001136019
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr7_+_149570004
|
5.964
|
NM_001100592
NM_145230
|
ATP6V0E2
|
ATPase, H+ transporting V0 subunit e2
|
|
chrX_+_47004619
|
5.874
|
|
RBM10
|
RNA binding motif protein 10
|
|
chr7_-_38313232
|
5.839
|
NM_001003799
NM_001003806
|
TARP
TRGV9
|
TCR gamma alternate reading frame protein
T cell receptor gamma variable 9
|
|
chr18_-_24442534
|
5.745
|
NM_004028
|
AQP4
|
aquaporin 4
|
|
chr2_-_85839147
|
5.731
|
|
C2orf68
|
chromosome 2 open reading frame 68
|
|
chr22_-_31688329
|
5.709
|
NM_001135911
NM_052880
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1
|
|
chr18_-_74728677
|
5.688
|
|
MBP
|
myelin basic protein
|
|
chr12_+_32112286
|
5.629
|
NM_018169
|
C12orf35
|
chromosome 12 open reading frame 35
|
|
chr8_+_85097099
|
5.590
|
NM_001100391
|
RALYL
|
RALY RNA binding protein-like
|
|
chr7_-_37393271
|
5.585
|
NM_001206482
|
ELMO1
|
engulfment and cell motility 1
|
|
chr15_-_82641705
|
5.553
|
NM_001164465
|
GOLGA6L10
|
golgin A6 family-like 10
|
|
chr14_+_101300806
|
5.496
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr12_+_10365415
|
5.486
|
NM_031412
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1
|
|
chr8_-_145653900
|
5.474
|
|
VPS28
|
vacuolar protein sorting 28 homolog (S. cerevisiae)
|
|
chr5_-_142783225
|
5.397
|
NM_000176
NM_001020825
NM_001024094
NM_001204258
NM_001204259
NM_001204260
NM_001204261
NM_001204262
NM_001204263
NM_001204264
NM_001204265
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr5_-_93447237
|
5.377
|
NM_001163417
NM_001163418
NM_032042
|
FAM172A
|
family with sequence similarity 172, member A
|
|
chr20_-_23062786
|
5.305
|
|
CD93
|
CD93 molecule
|
|
chr9_-_4742042
|
5.297
|
NM_001199853
|
AK3
|
adenylate kinase 3
|
|
chr14_-_60097314
|
5.283
|
|
RTN1
|
reticulon 1
|
|
chr4_-_170679027
|
5.234
|
NM_017867
|
C4orf27
|
chromosome 4 open reading frame 27
|
|
chr8_-_145653882
|
5.215
|
|
VPS28
|
vacuolar protein sorting 28 homolog (S. cerevisiae)
|
|
chr1_+_84543629
|
5.077
|
NM_002731
NM_207578
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr2_-_85839163
|
5.042
|
|
C2orf68
|
chromosome 2 open reading frame 68
|
|
chr15_-_83018159
|
5.020
|
NM_001164465
|
LOC643696
GOLGA6L10
|
uncharacterized LOC643696
golgin A6 family-like 10
|
|
chr3_-_47621729
|
4.845
|
NM_001206942
NM_001206945
|
CSPG5
|
chondroitin sulfate proteoglycan 5 (neuroglycan C)
|
|
chr8_-_145653912
|
4.818
|
NM_016208
NM_183057
|
VPS28
|
vacuolar protein sorting 28 homolog (S. cerevisiae)
|
|
chr7_-_99756329
|
4.812
|
|
C7orf43
|
chromosome 7 open reading frame 43
|
|
chrX_+_47004693
|
4.797
|
|
RBM10
|
RNA binding motif protein 10
|
|
chr6_-_90062450
|
4.714
|
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1, U
|
|
chr9_-_91793375
|
4.712
|
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr4_+_40198526
|
4.670
|
NM_004310
|
RHOH
|
ras homolog gene family, member H
|
|
chr7_+_80267782
|
4.636
|
NM_000072
NM_001001548
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr8_-_134309448
|
4.519
|
NM_001135242
NM_006096
|
NDRG1
|
N-myc downstream regulated 1
|
|
chr21_+_38791206
|
4.517
|
NM_130436
|
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A
|
|
chr18_-_53255361
|
4.496
|
|
TCF4
|
transcription factor 4
|
|
chr19_+_7504573
|
4.480
|
NM_001130955
|
ARHGEF18
|
Rho/Rac guanine nucleotide exchange factor (GEF) 18
|
|
chr4_+_88896899
|
4.475
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr3_-_33759540
|
4.427
|
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr5_+_36606667
|
4.419
|
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr3_-_142166782
|
4.363
|
NM_001042604
NM_019001
|
XRN1
|
5'-3' exoribonuclease 1
|
|
chr15_+_32933869
|
4.269
|
NM_001144757
NM_003020
|
SCG5
|
secretogranin V (7B2 protein)
|
|
chr1_-_154578775
|
4.266
|
NM_001193495
|
ADAR
|
adenosine deaminase, RNA-specific
|
|
chr4_-_174320616
|
4.213
|
NM_007281
|
SCRG1
|
stimulator of chondrogenesis 1
|
|
chr13_-_29292961
|
4.197
|
NM_001135919
NM_181785
|
SLC46A3
|
solute carrier family 46, member 3
|
|
chrY_+_2709596
|
4.185
|
NM_001008
|
RPS4Y1
|
ribosomal protein S4, Y-linked 1
|
|
chr19_+_17378264
|
4.085
|
|
BABAM1
|
BRISC and BRCA1 A complex member 1
|
|
chr16_-_28608367
|
4.082
|
NM_001054
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2
|
|
chr12_+_123717843
|
4.082
|
NM_152269
NM_001194995
|
C12orf65
|
chromosome 12 open reading frame 65
|
|
chr17_-_29648631
|
4.060
|
NM_001003927
NM_014210
|
EVI2A
|
ecotropic viral integration site 2A
|
|
chr2_-_136875629
|
4.014
|
NM_003467
|
CXCR4
|
chemokine (C-X-C motif) receptor 4
|
|
chr22_-_45608232
|
3.902
|
|
KIAA0930
|
KIAA0930
|
|
chr1_+_151032866
|
3.887
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr1_-_154928595
|
3.883
|
|
PBXIP1
|
pre-B-cell leukemia homeobox interacting protein 1
|
|
chr1_+_87794576
|
3.847
|
|
LMO4
|
LIM domain only 4
|
|
chr3_-_17741511
|
3.806
|
NM_001134381
|
TBC1D5
|
TBC1 domain family, member 5
|
|
chrY_+_2709630
|
3.802
|
|
RPS4Y1
|
ribosomal protein S4, Y-linked 1
|
|
chr11_+_66406087
|
3.794
|
NM_001198843
NM_001198844
NM_002896
|
RBM4
|
RNA binding motif protein 4
|
|
chr9_-_136933552
|
3.730
|
|
BRD3
|
bromodomain containing 3
|
|
chr12_+_123717458
|
3.701
|
|
C12orf65
|
chromosome 12 open reading frame 65
|
|
chr22_-_17680485
|
3.697
|
NM_177405
|
CECR1
|
cat eye syndrome chromosome region, candidate 1
|
|
chr3_+_42530790
|
3.685
|
NM_001251882
|
VIPR1
|
vasoactive intestinal peptide receptor 1
|
|
chr2_-_122406994
|
3.613
|
NM_001142273
NM_001142274
NM_001207051
NM_015282
|
CLASP1
|
cytoplasmic linker associated protein 1
|
|
chr19_+_55105046
|
3.601
|
NM_006863
|
LILRA1
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 1
|
|
chr22_-_38881529
|
3.590
|
|
DDX17
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 17
|
|
chr4_+_41258895
|
3.531
|
NM_004181
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chrX_-_6145887
|
3.437
|
NM_020742
|
NLGN4X
|
neuroligin 4, X-linked
|
|
chr3_+_111717585
|
3.424
|
NM_001008272
NM_013259
|
TAGLN3
|
transgelin 3
|
|
chr17_-_47841468
|
3.380
|
|
FAM117A
|
family with sequence similarity 117, member A
|
|
chr14_-_95623665
|
3.366
|
NM_177438
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr10_+_91152321
|
3.360
|
NM_001548
|
IFIT1
|
interferon-induced protein with tetratricopeptide repeats 1
|
|
chr2_+_99797632
|
3.351
|
|
MRPL30
|
mitochondrial ribosomal protein L30
|
|
chr5_+_161275897
|
3.350
|
NM_001127646
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr7_-_55930444
|
3.348
|
NM_207366
|
SEPT14
|
septin 14
|
|
chr3_+_122296432
|
3.272
|
NM_001113523
|
PARP15
|
poly (ADP-ribose) polymerase family, member 15
|
|
chr1_+_26644380
|
3.267
|
NM_001803
|
CD52
|
CD52 molecule
|
|
chr8_-_38854001
|
3.265
|
|
TM2D2
|
TM2 domain containing 2
|
|
chr11_+_124609841
|
3.263
|
|
NRGN
|
neurogranin (protein kinase C substrate, RC3)
|
|
chr1_-_177133950
|
3.250
|
NM_004319
NM_207108
|
ASTN1
|
astrotactin 1
|
|
chr4_+_492988
|
3.246
|
NM_001127178
NM_017733
|
PIGG
|
phosphatidylinositol glycan anchor biosynthesis, class G
|
|
chr10_-_75571521
|
3.224
|
NM_003635
|
NDST2
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2
|
|
chr3_-_17782398
|
3.221
|
NM_014744
|
TBC1D5
|
TBC1 domain family, member 5
|
|
chr12_-_10542616
|
3.185
|
NM_007360
|
KLRK1
|
killer cell lectin-like receptor subfamily K, member 1
|
|
chr1_+_231376918
|
3.173
|
NM_014236
|
GNPAT
|
glyceronephosphate O-acyltransferase
|
|
chr2_-_87018802
|
3.159
|
NM_001768
NM_171827
|
CD8A
|
CD8a molecule
|
|
chr10_+_6186758
|
3.155
|
NM_001145443
|
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
|
|
chr14_+_21249209
|
3.142
|
NM_005615
|
RNASE6
|
ribonuclease, RNase A family, k6
|
|
chr6_+_29910246
|
3.127
|
NM_001242758
NM_002116
|
HLA-A
|
major histocompatibility complex, class I, A
|
|
chr1_-_154928564
|
3.127
|
NM_020524
|
PBXIP1
|
pre-B-cell leukemia homeobox interacting protein 1
|
|
chr19_-_10420220
|
3.127
|
NM_001103167
|
ZGLP1
|
zinc finger, GATA-like protein 1
|
|
chr5_+_162930230
|
3.117
|
NM_182796
|
MAT2B
|
methionine adenosyltransferase II, beta
|
|
chr1_-_31230594
|
3.088
|
|
LAPTM5
|
lysosomal protein transmembrane 5
|
|
chr8_+_26435339
|
3.066
|
NM_001386
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr8_-_38854025
|
3.036
|
NM_001024380
NM_001024381
NM_031940
NM_078473
|
TM2D2
|
TM2 domain containing 2
|
|
chr17_+_53344966
|
3.028
|
|
HLF
|
hepatic leukemia factor
|
|
chr16_+_50731049
|
3.017
|
NM_022162
|
NOD2
|
nucleotide-binding oligomerization domain containing 2
|
|
chr6_+_33043702
|
2.977
|
NM_002121
|
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1
|
|
chr11_+_28131830
|
2.965
|
|
METTL15
|
methyltransferase like 15
|
|
chr3_+_9975513
|
2.964
|
NM_015513
NM_001031717
NM_001077415
|
CRELD1
|
cysteine-rich with EGF-like domains 1
|
|
chr11_+_34460421
|
2.956
|
NM_001752
|
CAT
|
catalase
|
|
chr1_-_1624145
|
2.945
|
|
SLC35E2B
|
solute carrier family 35, member E2B
|
|
chr1_+_231376959
|
2.932
|
|
GNPAT
|
glyceronephosphate O-acyltransferase
|
|
chr17_-_1412995
|
2.907
|
|
INPP5K
|
inositol polyphosphate-5-phosphatase K
|
|
chr1_+_26644458
|
2.900
|
|
CD52
|
CD52 molecule
|