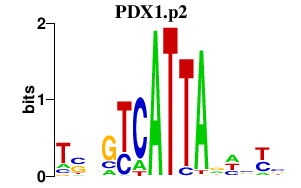
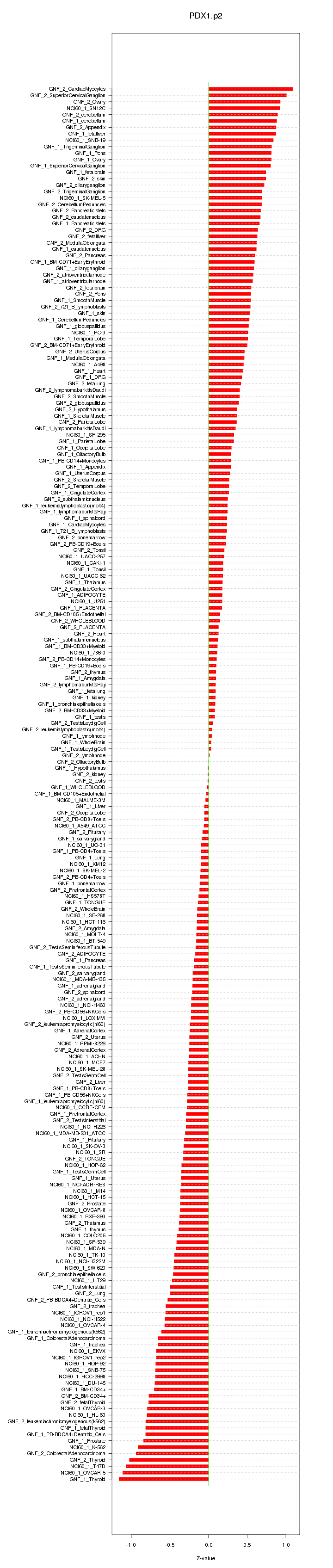
|
chr4_-_176733413
|
11.963
|
|
GPM6A
|
glycoprotein M6A
|
|
chr1_+_159175048
|
7.614
|
NM_001122951
|
DARC
|
Duffy blood group, chemokine receptor
|
|
chrX_+_110366304
|
5.893
|
NM_001128168
NM_001128172
NM_001128173
|
PAK3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr4_-_176733901
|
4.530
|
|
GPM6A
|
glycoprotein M6A
|
|
chr17_+_38119225
|
3.767
|
NM_178171
|
GSDMA
|
gasdermin A
|
|
chr7_-_44228880
|
3.680
|
NM_000162
|
GCK
|
glucokinase (hexokinase 4)
|
|
chr17_+_7482980
|
3.627
|
|
CD68
|
CD68 molecule
|
|
chr17_+_7482938
|
3.616
|
|
CD68
|
CD68 molecule
|
|
chr1_+_63063186
|
3.541
|
NM_014495
|
ANGPTL3
|
angiopoietin-like 3
|
|
chr4_+_86396251
|
3.360
|
NM_001025616
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr8_-_27115935
|
3.131
|
|
STMN4
|
stathmin-like 4
|
|
chr6_-_133035180
|
3.063
|
NM_004666
|
VNN1
|
vanin 1
|
|
chr16_+_70557688
|
3.022
|
NM_012426
|
SF3B3
|
splicing factor 3b, subunit 3, 130kDa
|
|
chr3_+_35722428
|
2.895
|
NM_198399
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr3_+_100238063
|
2.813
|
|
TMEM45A
|
transmembrane protein 45A
|
|
chr2_-_166060552
|
2.753
|
NM_001081676
NM_001081677
NM_006922
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit
|
|
chr3_+_115342150
|
2.648
|
NM_001130064
NM_002045
|
GAP43
|
growth associated protein 43
|
|
chr8_-_17579729
|
2.330
|
NM_001001931
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr6_-_31107110
|
2.271
|
NM_014069
|
PSORS1C2
|
psoriasis susceptibility 1 candidate 2
|
|
chr16_+_19078916
|
2.170
|
NM_016138
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr4_+_88720701
|
2.139
|
NM_004967
|
IBSP
|
integrin-binding sialoprotein
|
|
chr4_+_88896801
|
2.109
|
NM_000582
NM_001040058
NM_001040060
NM_001251829
NM_001251830
|
SPP1
|
secreted phosphoprotein 1
|
|
chr19_-_7766978
|
2.066
|
NM_001220500
NM_002002
|
FCER2
|
Fc fragment of IgE, low affinity II, receptor for (CD23)
|
|
chr12_-_54020081
|
2.016
|
NM_001130060
NM_001206682
NM_006856
|
ATF7
|
activating transcription factor 7
|
|
chr6_-_55740387
|
2.002
|
|
BMP5
|
bone morphogenetic protein 5
|
|
chr16_+_19078958
|
1.792
|
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr11_-_62457370
|
1.780
|
NM_203422
|
LRRN4CL
|
LRRN4 C-terminal like
|
|
chr17_+_7482804
|
1.710
|
NM_001251
NM_001040059
|
CD68
|
CD68 molecule
|
|
chr14_+_101295409
|
1.696
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr1_-_20250051
|
1.688
|
NM_014589
|
PLA2G2E
|
phospholipase A2, group IIE
|
|
chr6_-_41254392
|
1.688
|
NM_001242589
NM_001242590
NM_018643
|
TREM1
|
triggering receptor expressed on myeloid cells 1
|
|
chr6_+_72922513
|
1.439
|
NM_001168407
NM_001168408
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr3_-_21792815
|
1.438
|
NM_024697
|
ZNF385D
|
zinc finger protein 385D
|
|
chr12_-_4488707
|
1.398
|
NM_020638
|
FGF23
|
fibroblast growth factor 23
|
|
chr6_+_33043702
|
1.379
|
NM_002121
|
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1
|
|
chr17_+_7960201
|
1.345
|
|
|
|
|
chr1_+_163038934
|
1.343
|
NM_001113381
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr3_+_46619075
|
1.333
|
NM_003212
|
TDGF1
|
teratocarcinoma-derived growth factor 1
|
|
chr12_-_91398744
|
1.270
|
|
EPYC
|
epiphycan
|
|
chr2_+_169921298
|
1.262
|
NM_005771
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr2_+_185463235
|
1.244
|
|
ZNF804A
|
zinc finger protein 804A
|
|
chr8_-_17580024
|
1.228
|
NM_001166393
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr6_-_55740358
|
1.197
|
NM_021073
|
BMP5
|
bone morphogenetic protein 5
|
|
chr1_+_163038564
|
1.173
|
NM_005613
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr3_+_52813936
|
1.097
|
NM_001166435
NM_001166436
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1
|
|
chr9_+_82187469
|
1.062
|
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chr2_-_158300517
|
1.037
|
NM_004288
|
CYTIP
|
cytohesin 1 interacting protein
|
|
chr17_-_39637351
|
1.014
|
NM_002280
|
KRT35
|
keratin 35
|
|
chr17_-_29641068
|
0.974
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr11_-_66445318
|
0.966
|
|
RBM4B
|
RNA binding motif protein 4B
|
|
chr1_+_36748164
|
0.927
|
|
THRAP3
|
thyroid hormone receptor associated protein 3
|
|
chr12_+_75784878
|
0.927
|
NM_152436
|
GLIPR1L2
|
GLI pathogenesis-related 1 like 2
|
|
chr10_+_115310589
|
0.923
|
NM_001177660
|
HABP2
|
hyaluronan binding protein 2
|
|
chr6_-_62995852
|
0.907
|
NM_152688
|
KHDRBS2
|
KH domain containing, RNA binding, signal transduction associated 2
|
|
chr19_-_14952688
|
0.887
|
NM_001005190
|
OR7A10
|
olfactory receptor, family 7, subfamily A, member 10
|
|
chr6_-_46825942
|
0.799
|
|
GPR116
|
G protein-coupled receptor 116
|
|
chr1_+_101003692
|
0.789
|
NM_022049
|
GPR88
|
G protein-coupled receptor 88
|
|
chr1_+_92632608
|
0.785
|
NM_015237
|
KIAA1107
|
KIAA1107
|
|
chr9_+_82187599
|
0.782
|
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chr8_-_20040612
|
0.761
|
NM_001135691
NM_001142324
NM_001142325
NM_003053
|
SLC18A1
|
solute carrier family 18 (vesicular monoamine), member 1
|
|
chr10_-_62332666
|
0.754
|
NM_001204404
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr3_-_167191808
|
0.742
|
NM_006217
|
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2
|
|
chr16_+_19079220
|
0.737
|
NM_001190983
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr12_+_4829751
|
0.729
|
NM_017417
|
GALNT8
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 8 (GalNAc-T8)
|
|
chr8_-_17555158
|
0.724
|
NM_020749
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr7_+_93535819
|
0.715
|
NM_021955
|
GNGT1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1
|
|
chr12_-_10962673
|
0.705
|
NM_023917
|
TAS2R9
|
taste receptor, type 2, member 9
|
|
chr4_+_187187342
|
0.701
|
|
F11
|
coagulation factor XI
|
|
chr2_+_169926084
|
0.682
|
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr3_-_114343791
|
0.681
|
NM_001164346
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr5_+_140201221
|
0.680
|
NM_018908
NM_031501
|
PCDHA5
|
protocadherin alpha 5
|
|
chr4_+_113970784
|
0.671
|
NM_001148
NM_020977
|
ANK2
|
ankyrin 2, neuronal
|
|
chr6_+_25754926
|
0.654
|
NM_005495
|
SLC17A4
|
solute carrier family 17 (sodium phosphate), member 4
|
|
chr4_+_74606222
|
0.632
|
NM_000584
|
IL8
|
interleukin 8
|
|
chr9_-_21228132
|
0.624
|
NM_021268
|
IFNA17
|
interferon, alpha 17
|
|
chr1_-_204135420
|
0.620
|
NM_000537
|
REN
|
renin
|
|
chr11_+_88911039
|
0.585
|
NM_000372
|
TYR
|
tyrosinase (oculocutaneous albinism IA)
|
|
chr16_+_8814567
|
0.580
|
NM_001127448
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr12_-_103311380
|
0.554
|
NM_000277
|
PAH
|
phenylalanine hydroxylase
|
|
chr21_-_31538970
|
0.552
|
NM_012131
|
CLDN17
|
claudin 17
|
|
chr6_+_35310399
|
0.539
|
|
PPARD
|
peroxisome proliferator-activated receptor delta
|
|
chr4_-_10023094
|
0.538
|
NM_020041
|
SLC2A9
|
solute carrier family 2 (facilitated glucose transporter), member 9
|
|
chr5_-_75919051
|
0.533
|
NM_004101
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2
|
|
chr1_+_186265404
|
0.530
|
NM_001127708
NM_001127709
NM_001127710
NM_005807
|
PRG4
|
proteoglycan 4
|
|
chr6_+_131571298
|
0.523
|
NM_004842
NM_138633
|
AKAP7
|
A kinase (PRKA) anchor protein 7
|
|
chr14_+_70918873
|
0.522
|
NM_003813
|
ADAM21
|
ADAM metallopeptidase domain 21
|
|
chr7_-_113559048
|
0.520
|
NM_002711
|
PPP1R3A
|
protein phosphatase 1, regulatory subunit 3A
|
|
chr1_-_16763887
|
0.509
|
NM_198546
|
SPATA21
|
spermatogenesis associated 21
|
|
chr12_+_119616594
|
0.503
|
NM_014365
|
HSPB8
|
heat shock 22kDa protein 8
|
|
chr6_+_72926144
|
0.503
|
NM_001168409
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr4_+_78432905
|
0.475
|
NM_006419
|
CXCL13
|
chemokine (C-X-C motif) ligand 13
|
|
chr7_+_156435670
|
0.463
|
NM_001184997
|
RNF32
|
ring finger protein 32
|
|
chr15_+_67841341
|
0.458
|
NM_001206804
|
MAP2K5
|
mitogen-activated protein kinase kinase 5
|
|
chr12_+_51318515
|
0.456
|
NM_014033
|
METTL7A
|
methyltransferase like 7A
|
|
chr1_-_161601251
|
0.449
|
NM_001244753
|
FCGR3B
|
Fc fragment of IgG, low affinity IIIb, receptor (CD16b)
|
|
chr8_+_32579350
|
0.448
|
NM_001159996
|
NRG1
|
neuregulin 1
|
|
chr19_-_14785626
|
0.443
|
NM_032571
|
EMR3
|
egf-like module containing, mucin-like, hormone receptor-like 3
|
|
chr1_+_13911966
|
0.434
|
NM_001006624
NM_001006625
|
PDPN
|
podoplanin
|
|
chr3_+_151531860
|
0.421
|
NM_001086
|
AADAC
|
arylacetamide deacetylase (esterase)
|
|
chr11_+_28131830
|
0.417
|
|
METTL15
|
methyltransferase like 15
|
|
chr1_+_17906941
|
0.411
|
NM_001011722
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like
|
|
chr6_-_43423785
|
0.402
|
NM_206539
|
DLK2
|
delta-like 2 homolog (Drosophila)
|
|
chr12_-_121454285
|
0.397
|
NM_022895
|
C12orf43
|
chromosome 12 open reading frame 43
|
|
chr17_-_29641087
|
0.394
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr3_+_148447966
|
0.392
|
NM_032049
|
AGTR1
|
angiotensin II receptor, type 1
|
|
chr3_+_52811601
|
0.390
|
NM_002215
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1
|
|
chr17_-_38859976
|
0.383
|
NM_019016
|
KRT24
|
keratin 24
|
|
chr1_-_161519817
|
0.369
|
NM_000569
NM_001127592
|
FCGR3A
|
Fc fragment of IgG, low affinity IIIa, receptor (CD16a)
|
|
chr17_-_29641094
|
0.366
|
NM_006495
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr10_+_32856650
|
0.347
|
NM_024688
|
C10orf68
|
chromosome 10 open reading frame 68
|
|
chr6_-_138539626
|
0.326
|
NM_021635
|
PBOV1
|
prostate and breast cancer overexpressed 1
|
|
chr3_-_47933997
|
0.325
|
|
MAP4
|
microtubule-associated protein 4
|
|
chr5_-_16509106
|
0.325
|
NM_019000
|
FAM134B
|
family with sequence similarity 134, member B
|
|
chr1_-_186417855
|
0.314
|
NM_022576
|
PDC
|
phosducin
|
|
chr9_-_21239975
|
0.313
|
NM_002172
|
IFNA14
|
interferon, alpha 14
|
|
chr3_+_11178778
|
0.312
|
NM_001098213
|
HRH1
|
histamine receptor H1
|
|
chr11_-_104905849
|
0.285
|
|
CASP1
|
caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase)
|
|
chr4_+_71457974
|
0.283
|
NM_016519
|
AMBN
|
ameloblastin (enamel matrix protein)
|
|
chr5_+_140227356
|
0.283
|
NM_014005
NM_031857
|
PCDHA9
|
protocadherin alpha 9
|
|
chr12_-_91398757
|
0.281
|
NM_004950
|
EPYC
|
epiphycan
|
|
chr16_-_70557430
|
0.280
|
NM_001195139
NM_015386
|
COG4
|
component of oligomeric golgi complex 4
|
|
chr12_-_121454210
|
0.280
|
|
C12orf43
|
chromosome 12 open reading frame 43
|
|
chr4_+_74281884
|
0.272
|
|
ALB
|
albumin
|
|
chr4_-_46996423
|
0.262
|
NM_000809
|
GABRA4
|
gamma-aminobutyric acid (GABA) A receptor, alpha 4
|
|
chr7_+_129269916
|
0.252
|
NM_001040110
|
NRF1
|
nuclear respiratory factor 1
|
|
chr11_+_123986103
|
0.251
|
NM_001130142
NM_014622
NM_198315
|
VWA5A
|
von Willebrand factor A domain containing 5A
|
|
chr5_+_140718353
|
0.249
|
NM_018915
NM_032009
|
PCDHGA2
|
protocadherin gamma subfamily A, 2
|
|
chrX_+_37639306
|
0.246
|
|
CYBB
|
cytochrome b-245, beta polypeptide
|
|
chr7_+_26400503
|
0.245
|
NM_001199837
|
SNX10
|
sorting nexin 10
|
|
chr10_+_70980058
|
0.237
|
NM_025130
|
HKDC1
|
hexokinase domain containing 1
|
|
chr16_+_29674282
|
0.232
|
NM_001030288
|
SPN
|
sialophorin
|
|
chr7_-_27166638
|
0.230
|
NM_153631
|
HOXA3
|
homeobox A3
|
|
chr6_-_143266283
|
0.209
|
NM_006734
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2
|
|
chr7_-_44580857
|
0.196
|
NM_001101648
NM_013389
|
NPC1L1
|
NPC1 (Niemann-Pick disease, type C1, gene)-like 1
|
|
chr16_+_28505920
|
0.193
|
NM_018690
|
APOBR
|
apolipoprotein B receptor
|
|
chr2_-_216259197
|
0.191
|
|
FN1
|
fibronectin 1
|
|
chr5_-_16738346
|
0.180
|
|
MYO10
|
myosin X
|
|
chr12_+_10460416
|
0.168
|
NM_002262
NM_007334
|
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1
|
|
chr1_+_57320442
|
0.163
|
NM_000562
|
C8A
|
complement component 8, alpha polypeptide
|
|
chr6_-_134498973
|
0.158
|
NM_001143677
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chrX_-_62780748
|
0.156
|
|
LOC92249
|
uncharacterized LOC92249
|
|
chr3_-_178969644
|
0.154
|
NM_171830
|
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3
|
|
chr6_-_32731266
|
0.149
|
NM_001198858
|
HLA-DQB2
|
major histocompatibility complex, class II, DQ beta 2
|
|
chr1_+_84609941
|
0.149
|
NM_182948
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr3_+_108541544
|
0.146
|
NM_016388
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1
|
|
chr18_+_616671
|
0.133
|
NM_199167
|
CLUL1
|
clusterin-like 1 (retinal)
|
|
chr6_+_112375277
|
0.122
|
NM_003880
NM_198239
|
WISP3
|
WNT1 inducible signaling pathway protein 3
|
|
chr20_+_31037447
|
0.122
|
|
|
|
|
chr1_-_21605984
|
0.102
|
NM_001113347
|
ECE1
|
endothelin converting enzyme 1
|
|
chr12_+_4699243
|
0.098
|
NM_003845
|
DYRK4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4
|
|
chr5_+_54398473
|
0.098
|
NM_006144
|
GZMA
|
granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3)
|
|
chrX_+_99899388
|
0.093
|
|
SRPX2
|
sushi-repeat containing protein, X-linked 2
|
|
chr17_-_7082654
|
0.081
|
NM_001197216
NM_001671
|
ASGR1
|
asialoglycoprotein receptor 1
|
|
chr7_+_80275967
|
0.077
|
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chrX_+_99899162
|
0.075
|
NM_014467
|
SRPX2
|
sushi-repeat containing protein, X-linked 2
|
|
chr9_+_136501484
|
0.073
|
NM_000787
|
DBH
|
dopamine beta-hydroxylase (dopamine beta-monooxygenase)
|
|
chr17_-_10372875
|
0.058
|
NM_017533
|
MYH4
|
myosin, heavy chain 4, skeletal muscle
|
|
chr6_+_167704802
|
0.058
|
NM_001143947
NM_018974
|
UNC93A
|
unc-93 homolog A (C. elegans)
|
|
chrX_+_49091926
|
0.057
|
NM_014008
|
CCDC22
|
coiled-coil domain containing 22
|
|
chr6_+_167536240
|
0.055
|
NM_031409
|
CCR6
|
chemokine (C-C motif) receptor 6
|
|
chr2_-_218808705
|
0.035
|
NM_022648
|
TNS1
|
tensin 1
|
|
chr3_-_114478117
|
0.033
|
NM_001164344
NM_001164345
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr5_+_140235594
|
0.033
|
NM_018901
NM_031859
NM_031860
|
PCDHA10
|
protocadherin alpha 10
|
|
chr7_-_87342569
|
0.008
|
NM_000927
|
ABCB1
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1
|
|
chr15_+_48498497
|
0.004
|
NM_000338
NM_001184832
|
SLC12A1
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 1
|