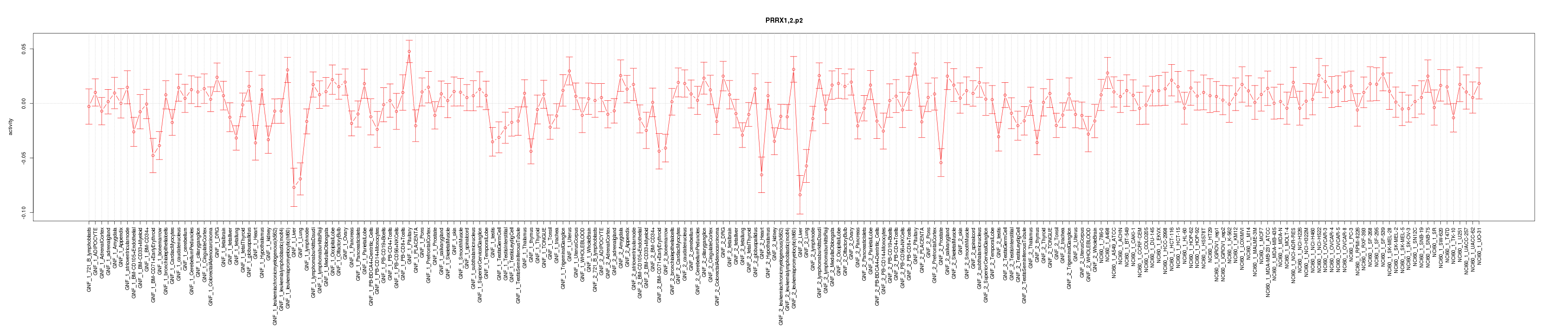
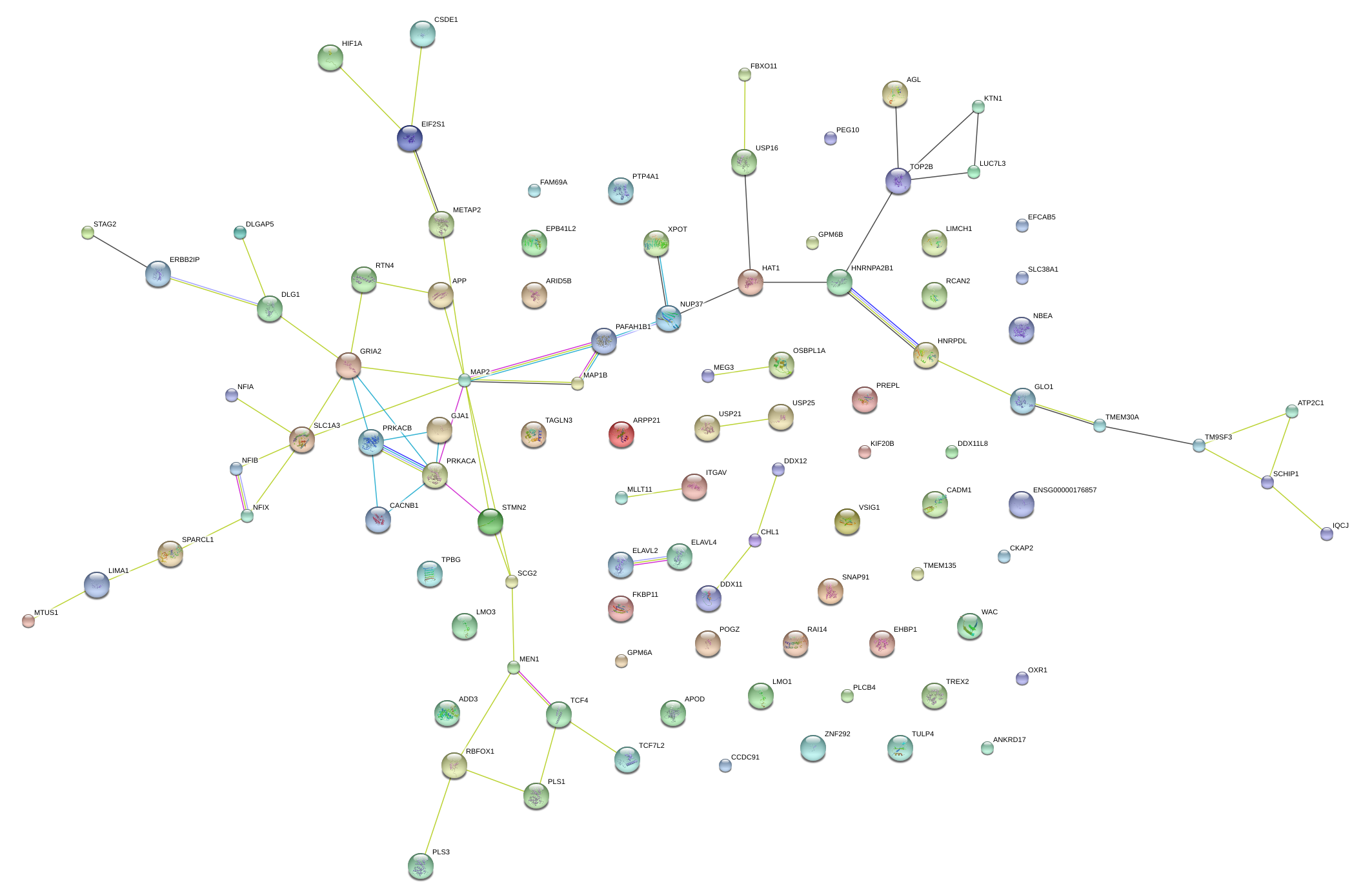
|
chr7_+_94297448
|
44.215
|
|
PEG10
|
paternally expressed 10
|
|
chr5_+_71502700
|
38.959
|
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr1_+_151032683
|
37.103
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr1_+_151032866
|
36.400
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr1_+_151032880
|
36.304
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr4_-_176733413
|
26.168
|
|
GPM6A
|
glycoprotein M6A
|
|
chr2_-_55199894
|
26.006
|
|
|
|
|
chr1_+_84630575
|
22.553
|
NM_001242860
NM_001242861
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chrX_-_13835012
|
21.858
|
|
GPM6B
|
glycoprotein M6B
|
|
chr3_+_158787040
|
21.134
|
NM_001197113
NM_001197114
NM_001042705
NM_001042706
NM_001197100
|
IQCJ-SCHIP1
IQCJ
|
IQCJ-SCHIP1 readthrough
IQ motif containing J
|
|
chr2_-_44586854
|
20.599
|
NM_001042385
NM_001042386
|
PREPL
|
prolyl endopeptidase-like
|
|
chr4_-_176733901
|
19.092
|
|
GPM6A
|
glycoprotein M6A
|
|
chr3_-_25683825
|
18.512
|
|
TOP2B
|
topoisomerase (DNA) II beta 180kDa
|
|
chr2_-_55237255
|
18.380
|
NM_007008
|
RTN4
|
reticulon 4
|
|
chrX_+_114827818
|
17.269
|
NM_001136025
NM_001172335
|
PLS3
|
plastin 3
|
|
chr1_+_84630374
|
17.049
|
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr3_+_159557745
|
16.989
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr17_-_1248346
|
16.812
|
|
PAFAH1B1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa)
|
|
chr4_+_158141735
|
16.503
|
NM_000826
NM_001083619
NM_001083620
|
GRIA2
|
glutamate receptor, ionotropic, AMPA 2
|
|
chr12_-_46633551
|
15.996
|
|
SLC38A1
|
solute carrier family 38, member 1
|
|
chr10_+_111765725
|
15.894
|
NM_019903
|
ADD3
|
adducin 3 (gamma)
|
|
chr6_-_75970566
|
15.500
|
|
TMEM30A
|
transmembrane protein 30A
|
|
chr3_+_159557649
|
15.237
|
NM_001197109
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr6_-_75965845
|
14.764
|
|
TMEM30A
|
transmembrane protein 30A
|
|
chr10_+_102123420
|
14.441
|
|
|
|
|
chr1_+_100316530
|
14.319
|
NM_000646
|
AGL
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase
|
|
chr21_+_30400229
|
14.222
|
|
USP16
|
ubiquitin specific peptidase 16
|
|
chr5_+_65372744
|
14.109
|
|
ERBB2IP
|
erbb2 interacting protein
|
|
chr1_+_84630014
|
13.755
|
NM_001242857
NM_001242858
NM_001242859
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr12_+_28410132
|
13.689
|
NM_018318
|
CCDC91
|
coiled-coil domain containing 91
|
|
chr1_+_84647342
|
13.655
|
NM_001242862
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr18_-_21891469
|
13.598
|
NM_001242508
|
OSBPL1A
|
oxysterol binding protein-like 1A
|
|
chr6_+_64231626
|
13.535
|
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr21_-_27253197
|
13.481
|
|
APP
|
amyloid beta (A4) precursor protein
|
|
chrX_+_123234462
|
13.466
|
|
STAG2
|
stromal antigen 2
|
|
chr3_+_361365
|
13.364
|
NM_001253388
|
CHL1
|
cell adhesion molecule with homology to L1CAM (close homolog of L1)
|
|
chr4_-_83347222
|
13.036
|
|
HNRPDL
|
heterogeneous nuclear ribonucleoprotein D-like
|
|
chr14_+_62164339
|
12.974
|
NM_001243084
|
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor)
|
|
chr8_+_80523351
|
12.655
|
|
STMN2
|
stathmin-like 2
|
|
chr10_+_28878666
|
12.648
|
|
WAC
|
WW domain containing adaptor with coiled-coil
|
|
chr1_-_93342706
|
12.538
|
NM_001252271
|
FAM69A
|
family with sequence similarity 69, member A
|
|
chr14_+_101295409
|
12.511
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr2_+_187454724
|
12.442
|
NM_001145000
NM_002210
|
ITGAV
|
integrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51)
|
|
chr4_-_74088773
|
12.300
|
|
ANKRD17
|
ankyrin repeat domain 17
|
|
chr17_+_28268622
|
11.975
|
NM_198529
|
EFCAB5
|
EF-hand calcium binding domain 5
|
|
chr3_-_196911001
|
11.866
|
NM_001204387
NM_001204388
|
DLG1
|
discs, large homolog 1 (Drosophila)
|
|
chr17_+_48821106
|
11.726
|
|
LUC7L3
|
LUC7-like 3 (S. cerevisiae)
|
|
chr4_-_88450243
|
11.712
|
|
SPARCL1
|
SPARC-like 1 (hevin)
|
|
chr10_+_63808968
|
11.644
|
NM_001244638
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chrX_+_107288199
|
11.586
|
NM_001170553
NM_182607
|
VSIG1
|
V-set and immunoglobulin domain containing 1
|
|
chr8_+_107738239
|
11.349
|
NM_001198534
NM_001198535
|
OXR1
|
oxidation resistance 1
|
|
chr9_-_14086901
|
11.279
|
|
NFIB
|
nuclear factor I/B
|
|
chr8_-_17555158
|
11.179
|
NM_020749
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr6_+_158733691
|
11.123
|
NM_001007466
NM_020245
|
TULP4
|
tubby like protein 4
|
|
chr18_-_53089722
|
10.895
|
NM_001243233
|
TCF4
|
transcription factor 4
|
|
chr2_-_224467095
|
10.795
|
|
SCG2
|
secretogranin II
|
|
chr6_+_83072922
|
10.691
|
NM_006670
|
TPBG
|
trophoblast glycoprotein
|
|
chr1_-_151414641
|
10.544
|
NM_207171
|
POGZ
|
pogo transposable element with ZNF domain
|
|
chr5_+_34687663
|
10.502
|
NM_001145523
|
RAI14
|
retinoic acid induced 14
|
|
chr8_+_80523376
|
10.454
|
|
STMN2
|
stathmin-like 2
|
|
chr5_+_36606667
|
10.450
|
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr4_-_176734181
|
10.437
|
NM_201591
|
GPM6A
|
glycoprotein M6A
|
|
chr8_+_80523304
|
10.368
|
|
STMN2
|
stathmin-like 2
|
|
chr8_+_80523339
|
10.107
|
|
STMN2
|
stathmin-like 2
|
|
chr2_-_44587812
|
10.004
|
NM_006036
|
PREPL
|
prolyl endopeptidase-like
|
|
chr3_+_158991035
|
9.949
|
NM_001197107
NM_001197108
NM_014575
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr12_-_102512277
|
9.945
|
NM_024057
|
NUP37
|
nucleoporin 37kDa
|
|
chr6_-_38670915
|
9.806
|
|
GLO1
|
glyoxalase I
|
|
chr14_+_67831484
|
9.748
|
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa
|
|
chr2_+_62934030
|
9.745
|
NM_001142614
|
EHBP1
|
EH domain binding protein 1
|
|
chr2_-_224467142
|
9.692
|
NM_003469
|
SCG2
|
secretogranin II
|
|
chr12_-_50595303
|
9.489
|
NM_001243775
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr12_-_50616425
|
9.455
|
NM_001113547
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr7_-_26232148
|
9.437
|
|
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1
|
|
chr13_+_53035227
|
9.418
|
|
CKAP2
|
cytoskeleton associated protein 2
|
|
chr12_+_64803741
|
9.371
|
|
XPOT
|
exportin, tRNA (nuclear export receptor for tRNAs)
|
|
chr11_+_87032428
|
9.348
|
|
TMEM135
|
transmembrane protein 135
|
|
chr5_+_36606456
|
9.201
|
NM_001166696
NM_004172
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr10_-_98290309
|
9.151
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr1_-_115268890
|
8.968
|
|
CSDE1
|
cold shock domain containing E1, RNA-binding
|
|
chr11_-_115088628
|
8.760
|
|
CADM1
|
cell adhesion molecule 1
|
|
chr6_-_46293628
|
8.723
|
NM_005822
|
RCAN2
|
regulator of calcineurin 2
|
|
chr6_-_131321868
|
8.691
|
NM_001252660
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2
|
|
chr1_+_50574601
|
8.669
|
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr3_-_195310770
|
8.609
|
|
APOD
|
apolipoprotein D
|
|
chr2_+_172821874
|
8.550
|
|
HAT1
|
histone acetyltransferase 1
|
|
chr3_+_130569368
|
8.516
|
NM_001199180
NM_001199181
NM_001199182
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1
|
|
chr3_+_111717585
|
8.510
|
NM_001008272
NM_013259
|
TAGLN3
|
transgelin 3
|
|
chr6_-_84419018
|
8.441
|
|
SNAP91
|
synaptosomal-associated protein, 91kDa homolog (mouse)
|
|
chr22_-_36236264
|
8.309
|
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr6_-_38670922
|
8.230
|
NM_006708
|
GLO1
|
glyoxalase I
|
|
chr2_-_224467049
|
8.202
|
|
SCG2
|
secretogranin II
|
|
chr6_-_38670904
|
8.164
|
|
GLO1
|
glyoxalase I
|
|
chr20_+_9288446
|
8.120
|
NM_000933
|
PLCB4
|
phospholipase C, beta 4
|
|
chr3_+_142342265
|
8.105
|
NM_002670
|
PLS1
|
plastin 1
|
|
chr2_-_48115846
|
8.104
|
NM_025133
|
FBXO11
|
F-box protein 11
|
|
chr4_+_41700736
|
8.069
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr3_+_35722428
|
8.039
|
NM_198399
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr6_-_38670882
|
8.021
|
|
GLO1
|
glyoxalase I
|
|
chr12_-_16759733
|
7.995
|
NM_001243610
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr6_+_121756744
|
7.972
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chrX_-_13835187
|
7.866
|
|
GPM6B
|
glycoprotein M6B
|
|
chr14_+_56078735
|
7.852
|
|
KTN1
|
kinectin 1 (kinesin receptor)
|
|
chr6_+_87969619
|
7.801
|
|
ZNF292
|
zinc finger protein 292
|
|
chr22_-_36236330
|
7.762
|
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr6_-_84419108
|
7.702
|
NM_001242793
NM_001242794
NM_014841
|
SNAP91
|
synaptosomal-associated protein, 91kDa homolog (mouse)
|
|
chr13_+_36050885
|
7.679
|
NM_001204197
|
NBEA
|
neurobeachin
|
|
chr1_+_50569581
|
7.600
|
NM_001144776
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr8_+_80523048
|
7.589
|
NM_001199214
NM_007029
|
STMN2
|
stathmin-like 2
|
|
chr22_-_36236421
|
7.565
|
NM_001031695
NM_001082576
NM_001082577
NM_014309
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr10_+_91498724
|
7.538
|
|
KIF20B
|
kinesin family member 20B
|
|
chr16_+_7382750
|
7.519
|
NM_145891
NM_145892
NM_145893
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr2_+_210444154
|
7.424
|
|
MAP2
|
microtubule-associated protein 2
|
|
chr1_-_103496768
|
7.421
|
|
COL11A1
|
collagen, type XI, alpha 1
|
|
chr14_-_20801427
|
7.386
|
NM_021178
NM_182849
|
CCNB1IP1
|
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase
|
|
chr12_-_16759430
|
7.386
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr3_+_35721115
|
7.383
|
NM_001025068
NM_001025069
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr12_-_16759939
|
7.301
|
NM_001243613
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr15_-_25650647
|
7.275
|
NM_130838
|
UBE3A
|
ubiquitin protein ligase E3A
|
|
chr2_-_200325200
|
7.169
|
|
SATB2
|
SATB homeobox 2
|
|
chr2_-_61764661
|
7.051
|
|
XPO1
|
exportin 1 (CRM1 homolog, yeast)
|
|
chr5_+_71403040
|
7.047
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr3_+_141043054
|
6.965
|
NM_001080412
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr15_+_32933869
|
6.950
|
NM_001144757
NM_003020
|
SCG5
|
secretogranin V (7B2 protein)
|
|
chr8_-_62602285
|
6.930
|
NM_001164750
NM_001164751
NM_001164752
NM_001164753
NM_020164
NM_032467
NM_032468
|
ASPH
|
aspartate beta-hydroxylase
|
|
chr7_-_7758237
|
6.796
|
NM_002947
|
RPA3
|
replication protein A3, 14kDa
|
|
chr15_-_56209305
|
6.776
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr2_+_183608329
|
6.769
|
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10
|
|
chr2_+_170662044
|
6.669
|
|
SSB
|
Sjogren syndrome antigen B (autoantigen La)
|
|
chrX_-_13835313
|
6.638
|
NM_001001995
NM_001001996
NM_005278
|
GPM6B
|
glycoprotein M6B
|
|
chr4_+_169013687
|
6.634
|
NM_007193
|
ANXA10
|
annexin A10
|
|
chr7_-_107880492
|
6.632
|
NM_001037132
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr9_-_114247024
|
6.587
|
NM_001080398
|
KIAA0368
|
KIAA0368
|
|
chrX_-_10535476
|
6.497
|
NM_001193278
NM_001193279
NM_001193280
NM_001193281
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr1_+_50574589
|
6.476
|
NM_001144774
NM_021952
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr11_-_73471136
|
6.467
|
NM_001243719
|
RAB6A
|
RAB6A, member RAS oncogene family
|
|
chr1_+_92632608
|
6.398
|
NM_015237
|
KIAA1107
|
KIAA1107
|
|
chr10_+_63661012
|
6.389
|
NM_032199
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr14_+_70236266
|
6.379
|
|
SRSF5
|
serine/arginine-rich splicing factor 5
|
|
chr12_-_16760935
|
6.336
|
NM_001001395
NM_001243609
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr2_-_225378320
|
6.291
|
|
CUL3
|
cullin 3
|
|
chr5_+_132432880
|
6.278
|
|
HSPA4
|
heat shock 70kDa protein 4
|
|
chr18_+_33234532
|
6.270
|
NM_020474
|
GALNT1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 (GalNAc-T1)
|
|
chr6_-_38670871
|
6.246
|
|
GLO1
|
glyoxalase I
|
|
chr14_+_61201450
|
6.220
|
NM_001177963
NM_002431
|
MNAT1
|
menage a trois homolog 1, cyclin H assembly factor (Xenopus laevis)
|
|
chr12_+_50869531
|
6.192
|
|
LARP4
|
La ribonucleoprotein domain family, member 4
|
|
chr8_-_110661007
|
6.148
|
NM_001099747
NM_001099748
NM_001099749
NM_001099750
NM_017786
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr13_-_24007822
|
6.141
|
NM_014363
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin)
|
|
chr6_-_79675672
|
6.137
|
|
PHIP
|
pleckstrin homology domain interacting protein
|
|
chr4_+_146031528
|
6.119
|
|
ABCE1
|
ATP-binding cassette, sub-family E (OABP), member 1
|
|
chr2_-_225370687
|
6.057
|
|
CUL3
|
cullin 3
|
|
chr4_+_41614725
|
5.945
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr4_+_88896899
|
5.933
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr12_-_56679729
|
5.900
|
|
CS
|
citrate synthase
|
|
chr5_+_36608430
|
5.879
|
NM_001166695
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr2_-_20527120
|
5.870
|
NM_015317
|
PUM2
|
pumilio homolog 2 (Drosophila)
|
|
chr4_+_88896838
|
5.868
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr3_+_115342302
|
5.864
|
|
GAP43
|
growth associated protein 43
|
|
chr12_-_54653369
|
5.842
|
NM_001127321
|
CBX5
|
chromobox homolog 5
|
|
chr7_+_112063131
|
5.841
|
NM_001007245
NM_001197080
|
IFRD1
|
interferon-related developmental regulator 1
|
|
chr21_-_27512668
|
5.826
|
NM_001136016
|
APP
|
amyloid beta (A4) precursor protein
|
|
chr8_+_66582107
|
5.815
|
NM_001145838
|
MTFR1
|
mitochondrial fission regulator 1
|
|
chrX_-_13835213
|
5.798
|
|
GPM6B
|
glycoprotein M6B
|
|
chr4_-_139163222
|
5.717
|
NM_014331
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11
|
|
chr2_+_102508333
|
5.681
|
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr2_+_102508377
|
5.674
|
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr2_-_20512016
|
5.641
|
|
PUM2
|
pumilio homolog 2 (Drosophila)
|
|
chr11_+_69467210
|
5.608
|
|
CCND1
|
cyclin D1
|
|
chr20_+_11898536
|
5.606
|
NM_014962
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr1_+_163038934
|
5.584
|
NM_001113381
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr1_+_115572414
|
5.576
|
NM_000549
|
TSHB
|
thyroid stimulating hormone, beta
|
|
chr3_-_195310985
|
5.565
|
NM_001647
|
APOD
|
apolipoprotein D
|
|
chr6_-_6007097
|
5.560
|
|
NRN1
|
neuritin 1
|
|
chr5_+_125758818
|
5.513
|
NM_001146320
NM_001146321
NM_023927
|
GRAMD3
|
GRAM domain containing 3
|
|
chr12_+_75784878
|
5.492
|
NM_152436
|
GLIPR1L2
|
GLI pathogenesis-related 1 like 2
|
|
chr4_+_166131170
|
5.486
|
NM_001161521
NM_001161522
|
KLHL2
|
kelch-like 2, Mayven (Drosophila)
|
|
chr13_-_36429798
|
5.481
|
NM_001195415
NM_001195416
NM_001195430
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr2_-_10942642
|
5.476
|
|
PDIA6
|
protein disulfide isomerase family A, member 6
|
|
chr6_-_86351156
|
5.411
|
NM_001159675
NM_001159676
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr1_+_67632168
|
5.385
|
NM_144701
|
IL23R
|
interleukin 23 receptor
|
|
chr12_-_71551564
|
5.381
|
|
TSPAN8
|
tetraspanin 8
|
|
chr6_+_64286348
|
5.297
|
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr8_-_110620220
|
5.229
|
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr1_+_15990300
|
5.171
|
|
|
|
|
chr11_+_43665772
|
5.164
|
|
|
|
|
chr9_-_14398981
|
5.147
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chr12_-_54653317
|
5.136
|
NM_001127322
|
CBX5
|
chromobox homolog 5
|
|
chr12_-_16759531
|
5.113
|
NM_018640
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr10_-_95241943
|
5.102
|
NM_013451
NM_133337
|
MYOF
|
myoferlin
|
|
chr5_+_125695787
|
5.085
|
NM_001146319
|
GRAMD3
|
GRAM domain containing 3
|
|
chr8_+_98703361
|
5.027
|
|
MTDH
|
metadherin
|
|
chr1_-_23886284
|
5.019
|
NM_002167
|
ID3
|
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein
|
|
chr11_-_17191287
|
4.914
|
NM_002645
|
PIK3C2A
|
phosphoinositide-3-kinase, class 2, alpha polypeptide
|
|
chr5_+_147795459
|
4.905
|
|
FBXO38
|
F-box protein 38
|
|
chr5_+_179132676
|
4.891
|
|
CANX
|
calnexin
|
|
chr15_+_66694216
|
4.878
|
|
MAP2K1
|
mitogen-activated protein kinase kinase 1
|
|
chr17_-_35656691
|
4.874
|
NM_198837
NM_198838
|
ACACA
|
acetyl-CoA carboxylase alpha
|
|
chr19_+_30863318
|
4.859
|
NM_014717
|
ZNF536
|
zinc finger protein 536
|
|
chr10_-_118680978
|
4.820
|
|
KIAA1598
|
KIAA1598
|
|
chr5_-_86703927
|
4.816
|
|
CCNH
|
cyclin H
|