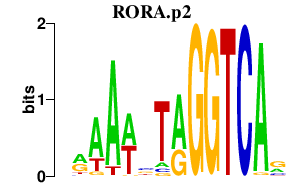
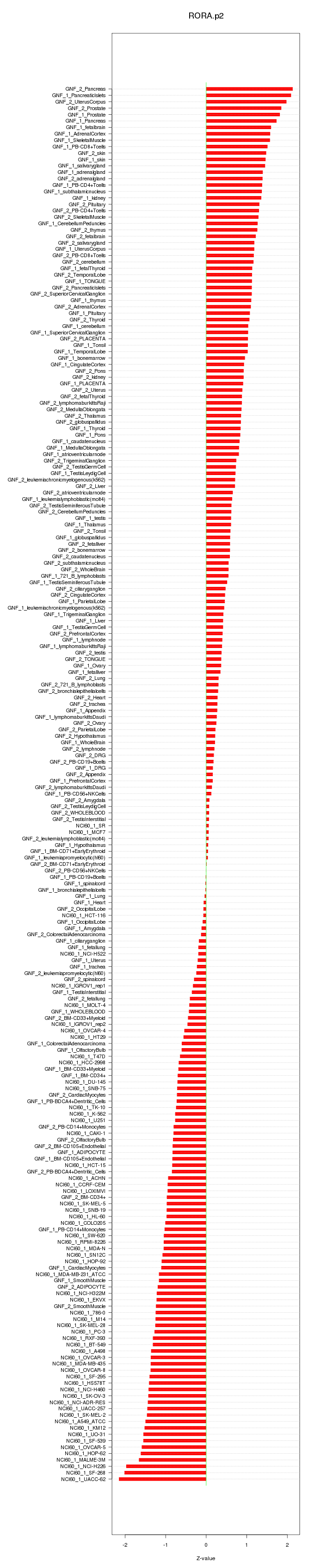
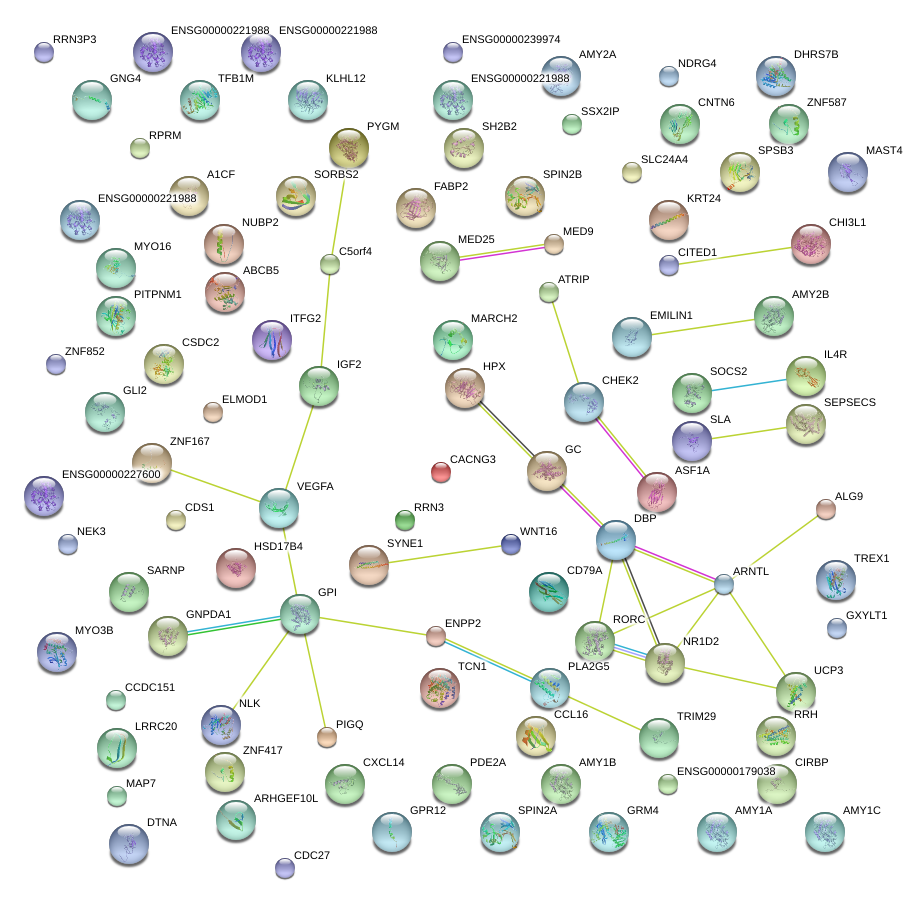
|
chr19_-_49140569
|
22.558
|
NM_001352
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein
|
|
chr10_-_52645384
|
17.731
|
|
A1CF
|
APOBEC1 complementation factor
|
|
chr12_-_42538432
|
16.620
|
NM_001099650
NM_173601
|
GXYLT1
|
glucoside xylosyltransferase 1
|
|
chr11_+_107461955
|
15.833
|
|
ELMOD1
|
ELMO/CED-12 domain containing 1
|
|
chr1_+_104198140
|
15.076
|
NM_001008221
|
AMY1A
|
amylase, alpha 1A (salivary)
|
|
chr1_-_235812994
|
14.679
|
NM_001098722
|
GNG4
|
guanine nucleotide binding protein (G protein), gamma 4
|
|
chr11_+_13299260
|
13.936
|
NM_001030272
NM_001030273
NM_001178
|
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like
|
|
chr4_-_186732208
|
13.464
|
NM_001145671
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr6_-_152623213
|
13.382
|
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr11_+_13299372
|
13.059
|
|
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like
|
|
chr11_+_13299376
|
12.766
|
|
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like
|
|
chr11_+_13299349
|
12.543
|
|
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like
|
|
chr11_+_107462073
|
12.252
|
|
ELMOD1
|
ELMO/CED-12 domain containing 1
|
|
chr16_-_1832571
|
12.110
|
|
SPSB3
|
splA/ryanodine receptor domain and SOCS box containing 3
|
|
chr16_-_1832576
|
11.735
|
NM_080861
|
SPSB3
|
splA/ryanodine receptor domain and SOCS box containing 3
|
|
chr5_-_134914968
|
11.733
|
NM_004887
|
CXCL14
|
chemokine (C-X-C motif) ligand 14
|
|
chr16_-_1832304
|
11.592
|
|
SPSB3
|
splA/ryanodine receptor domain and SOCS box containing 3
|
|
chr19_+_1269244
|
11.208
|
NM_001280
|
CIRBP
|
cold inducible RNA binding protein
|
|
chr11_+_13299391
|
10.904
|
|
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like
|
|
chr11_-_120008801
|
10.799
|
|
TRIM29
|
tripartite motif containing 29
|
|
chr8_-_134072404
|
9.970
|
NM_006748
|
SLA
|
Src-like-adaptor
|
|
chr11_+_13299329
|
9.819
|
|
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like
|
|
chr6_-_136847743
|
9.176
|
NM_001198614
NM_001198618
NM_001198619
|
MAP7
|
microtubule-associated protein 7
|
|
chr12_-_56211503
|
8.861
|
|
SARNP
|
SAP domain containing ribonucleoprotein
|
|
chr7_+_101928417
|
7.758
|
|
SH2B2
|
SH2B adaptor protein 2
|
|
chr11_-_64527369
|
7.739
|
|
PYGM
|
phosphorylase, glycogen, muscle
|
|
chr1_+_17846818
|
7.436
|
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like
|
|
chr6_+_43738757
|
7.251
|
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr2_-_16081703
|
6.812
|
|
MYCNOS
|
MYCN opposite strand/antisense RNA (non-protein coding)
|
|
chr7_+_101928390
|
6.801
|
NM_020979
|
SH2B2
|
SH2B adaptor protein 2
|
|
chr2_+_121554866
|
6.528
|
NM_005270
|
GLI2
|
GLI family zinc finger 2
|
|
chr17_-_34308455
|
6.370
|
NM_004590
|
CCL16
|
chemokine (C-C motif) ligand 16
|
|
chr3_+_44596596
|
6.200
|
NM_018651
NM_025169
|
ZNF167
|
zinc finger protein 167
|
|
chr16_-_21831730
|
6.095
|
|
RRN3P1
|
RNA polymerase I transcription factor homolog (S. cerevisiae) pseudogene 1
|
|
chr16_+_58534045
|
5.977
|
NM_001242833
NM_001242834
NM_001242835
NM_001242836
|
NDRG4
|
NDRG family member 4
|
|
chr11_-_2160199
|
5.962
|
NM_000612
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr1_-_203155867
|
5.921
|
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39)
|
|
chrX_-_71527000
|
5.884
|
NM_001144886
|
CITED1
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1
|
|
chr1_-_203155910
|
5.835
|
NM_001276
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39)
|
|
chr11_+_107461807
|
5.766
|
NM_001130037
NM_018712
|
ELMOD1
|
ELMO/CED-12 domain containing 1
|
|
chr1_+_104198301
|
5.670
|
NM_004038
NM_001008218
NM_001008219
|
AMY1A
AMY1B
AMY1C
|
amylase, alpha 1A (salivary)
amylase, alpha 1B (salivary)
amylase, alpha 1C (salivary)
|
|
chr1_-_104239071
|
5.651
|
NM_001008221
|
AMY1A
|
amylase, alpha 1A (salivary)
|
|
chr10_-_52645430
|
5.650
|
NM_001198818
NM_001198819
NM_001198820
NM_014576
NM_138932
NM_138933
|
A1CF
|
APOBEC1 complementation factor
|
|
chr2_-_154335181
|
5.505
|
NM_019845
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator candidate
|
|
chr3_+_1134619
|
5.201
|
NM_014461
|
CNTN6
|
contactin 6
|
|
chr13_-_27334777
|
5.167
|
NM_005288
|
GPR12
|
G protein-coupled receptor 12
|
|
chr11_-_67271839
|
4.894
|
|
PITPNM1
|
phosphatidylinositol transfer protein, membrane-associated 1
|
|
chr4_-_120243257
|
4.578
|
NM_000134
|
FABP2
|
fatty acid binding protein 2, intestinal
|
|
chr12_+_93963597
|
4.571
|
NM_003877
|
SOCS2
|
suppressor of cytokine signaling 2
|
|
chr11_-_2159784
|
4.549
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr16_+_27325269
|
4.539
|
|
IL4R
|
interleukin 4 receptor
|
|
chr5_-_154230157
|
4.467
|
|
C5orf4
|
chromosome 5 open reading frame 4
|
|
chr17_-_38859976
|
4.441
|
NM_019016
|
KRT24
|
keratin 24
|
|
chr5_+_66124603
|
4.284
|
NM_015183
|
MAST4
|
microtubule associated serine/threonine kinase family member 4
|
|
chr8_-_120651019
|
4.048
|
NM_001040092
NM_001130863
NM_006209
|
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2
|
|
chr10_-_72141347
|
3.773
|
NM_207119
|
LRRC20
|
leucine rich repeat containing 20
|
|
chr19_+_50321503
|
3.736
|
NM_030973
|
MED25
|
mediator complex subunit 25
|
|
chr1_-_85155885
|
3.731
|
NM_001166295
NM_001166417
NM_001166293
NM_001166294
NM_014021
|
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein
|
|
chr2_+_27301434
|
3.701
|
NM_007046
|
EMILIN1
|
elastin microfibril interfacer 1
|
|
chr7_+_120969089
|
3.595
|
NM_057168
|
WNT16
|
wingless-type MMTV integration site family, member 16
|
|
chr22_+_41957011
|
3.568
|
NM_014460
|
CSDC2
|
cold shock domain containing C2, RNA binding
|
|
chr11_-_6462129
|
3.458
|
NM_000613
|
HPX
|
hemopexin
|
|
chr16_+_1832928
|
3.343
|
NM_012225
|
NUBP2
|
nucleotide binding protein 2
|
|
chr16_+_24266873
|
3.222
|
NM_006539
|
CACNG3
|
calcium channel, voltage-dependent, gamma subunit 3
|
|
chr13_-_52733995
|
3.212
|
NM_001146099
NM_002498
|
NEK3
|
NIMA (never in mitosis gene a)-related kinase 3
|
|
chr7_+_20655244
|
3.181
|
NM_001163941
|
ABCB5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5
|
|
chr6_-_155635562
|
3.067
|
|
TFB1M
|
transcription factor B1, mitochondrial
|
|
chr1_+_20396652
|
3.030
|
NM_000929
|
PLA2G5
|
phospholipase A2, group V
|
|
chr18_+_32073245
|
2.996
|
NM_001198945
NM_001392
NM_032975
NM_032979
|
DTNA
|
dystrobrevin, alpha
|
|
chr16_-_15188089
|
2.992
|
NM_018427
|
RRN3
|
RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae)
|
|
chr11_-_120008738
|
2.968
|
|
TRIM29
|
tripartite motif containing 29
|
|
chr14_+_92788924
|
2.867
|
NM_153648
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr6_+_32121228
|
2.859
|
NM_138717
|
PPT2
|
palmitoyl-protein thioesterase 2
|
|
chr1_-_151798552
|
2.855
|
NM_001001523
|
RORC
|
RAR-related orphan receptor C
|
|
chr4_+_110749149
|
2.724
|
NM_006583
|
RRH
|
retinal pigment epithelium-derived rhodopsin homolog
|
|
chrX_-_57147850
|
2.542
|
NM_001006681
NM_001006682
NM_001006683
|
SPIN2B
|
spindlin family, member 2B
|
|
chr19_+_58361219
|
2.524
|
|
ZNF587
|
zinc finger protein 587
|
|
chr6_-_34101272
|
2.504
|
NM_000841
|
GRM4
|
glutamate receptor, metabotropic 4
|
|
chr4_+_85504056
|
2.368
|
NM_001263
|
CDS1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1
|
|
chr3_+_23987514
|
2.340
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr1_-_202896315
|
2.216
|
|
KLHL12
|
kelch-like 12 (Drosophila)
|
|
chr3_+_48488180
|
2.212
|
NM_032166
NM_130384
|
TREX1
ATRIP
|
three prime repair exonuclease 1
ATR interacting protein
|
|
chr2_+_139259476
|
2.175
|
|
|
|
|
chr19_+_42381189
|
2.173
|
NM_001783
NM_021601
|
CD79A
|
CD79a molecule, immunoglobulin-associated alpha
|
|
chr11_-_72380107
|
2.170
|
NM_001146209
|
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated
|
|
chr12_+_2921786
|
2.159
|
NM_018463
|
ITFG2
|
integrin alpha FG-GAP repeat containing 2
|
|
chr13_+_109248499
|
2.148
|
NM_015011
|
MYO16
|
myosin XVI
|
|
chr2_+_171034654
|
2.115
|
NM_001083615
NM_001171642
NM_138995
|
MYO3B
|
myosin IIIB
|
|
chr11_-_73720281
|
1.996
|
NM_003356
NM_022803
|
UCP3
|
uncoupling protein 3 (mitochondrial, proton carrier)
|
|
chr19_+_8478181
|
1.991
|
NM_001005415
NM_001005416
NM_016496
|
MARCH2
|
membrane-associated ring finger (C3HC4) 2
|
|
chr17_+_26369617
|
1.924
|
NM_016231
|
NLK
|
nemo-like kinase
|
|
chr10_+_51549552
|
1.921
|
NM_002443
NM_138634
|
MSMB
|
microseminoprotein, beta-
|
|
chr17_+_40688264
|
1.907
|
|
NAGLU
|
N-acetylglucosaminidase, alpha
|
|
chr1_-_156786500
|
1.906
|
NM_001161441
NM_001161442
NM_001161444
NM_003975
|
SH2D2A
|
SH2 domain containing 2A
|
|
chr17_+_7554711
|
1.864
|
|
ATP1B2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide
|
|
chr1_+_67632168
|
1.857
|
NM_144701
|
IL23R
|
interleukin 23 receptor
|
|
chr2_-_228497801
|
1.828
|
NM_001162483
NM_020161
|
C2orf83
|
chromosome 2 open reading frame 83
|
|
chr7_+_129008044
|
1.816
|
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr6_-_111927082
|
1.806
|
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chr10_-_72362513
|
1.796
|
NM_001083116
NM_005041
|
PRF1
|
perforin 1 (pore forming protein)
|
|
chr10_+_18041217
|
1.783
|
NM_001098844
|
TMEM236
|
transmembrane protein 236
|
|
chr1_+_154966061
|
1.778
|
NM_018655
|
LENEP
|
lens epithelial protein
|
|
chr19_+_16059817
|
1.757
|
NM_001004465
|
OR10H4
|
olfactory receptor, family 10, subfamily H, member 4
|
|
chr7_+_97736118
|
1.751
|
NM_014916
|
LMTK2
|
lemur tyrosine kinase 2
|
|
chr19_+_57791794
|
1.743
|
NM_006635
|
ZNF460
|
zinc finger protein 460
|
|
chr1_-_173176314
|
1.726
|
NM_003326
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4
|
|
chrX_-_102774416
|
1.714
|
NM_080879
|
RAB40A
|
RAB40A, member RAS oncogene family
|
|
chr12_-_48119224
|
1.709
|
NM_001172439
NM_001172440
NM_006025
|
ENDOU
|
endonuclease, polyU-specific
|
|
chr17_+_73043319
|
1.707
|
|
KCTD2
|
potassium channel tetramerisation domain containing 2
|
|
chr11_-_2159472
|
1.674
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chrX_-_62648209
|
1.614
|
|
LOC92249
|
uncharacterized LOC92249
|
|
chrX_-_32173585
|
1.607
|
NM_004013
NM_004020
NM_004021
NM_004022
NM_004023
|
DMD
|
dystrophin
|
|
chr10_-_72362441
|
1.607
|
|
PRF1
|
perforin 1 (pore forming protein)
|
|
chr11_-_102651321
|
1.606
|
NM_002425
|
MMP10
|
matrix metallopeptidase 10 (stromelysin 2)
|
|
chr1_-_203155827
|
1.573
|
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39)
|
|
chr18_+_32073343
|
1.573
|
|
DTNA
|
dystrobrevin, alpha
|
|
chr14_-_75389144
|
1.569
|
NM_031464
|
RPS6KL1
|
ribosomal protein S6 kinase-like 1
|
|
chr17_-_2304217
|
1.555
|
NM_020310
|
MNT
|
MAX binding protein
|
|
chr11_-_124806302
|
1.553
|
NM_152722
|
HEPACAM
|
hepatic and glial cell adhesion molecule
|
|
chr4_+_156824846
|
1.512
|
NM_005651
|
TDO2
|
tryptophan 2,3-dioxygenase
|
|
chr3_+_151531860
|
1.475
|
NM_001086
|
AADAC
|
arylacetamide deacetylase (esterase)
|
|
chr11_-_129062092
|
1.474
|
NM_001142685
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr15_+_45422130
|
1.473
|
NM_017434
NM_175940
|
DUOX1
|
dual oxidase 1
|
|
chr13_-_103719195
|
1.440
|
NM_000452
|
SLC10A2
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 2
|
|
chr1_+_111833473
|
1.439
|
NM_021797
NM_201653
|
CHIA
|
chitinase, acidic
|
|
chr11_+_131780537
|
1.425
|
NM_001144058
NM_001144059
NM_016522
|
NTM
|
neurotrimin
|
|
chr15_-_77176191
|
1.415
|
NM_020843
|
SCAPER
|
S-phase cyclin A-associated protein in the ER
|
|
chr5_-_137071703
|
1.400
|
NM_017415
|
KLHL3
|
kelch-like 3 (Drosophila)
|
|
chr18_-_3874688
|
1.391
|
NM_001242762
NM_001242763
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1
|
|
chr6_-_30181197
|
1.380
|
NM_001242783
NM_003449
|
TRIM26
|
tripartite motif containing 26
|
|
chr16_-_90096308
|
1.357
|
NM_001214
|
C16orf3
|
chromosome 16 open reading frame 3
|
|
chr1_+_157963062
|
1.334
|
NM_018240
|
KIRREL
|
kin of IRRE like (Drosophila)
|
|
chr12_+_56623819
|
1.251
|
NM_173596
|
SLC39A5
|
solute carrier family 39 (metal ion transporter), member 5
|
|
chr1_-_120311466
|
1.248
|
NM_001166107
NM_005518
|
HMGCS2
|
3-hydroxy-3-methylglutaryl-CoA synthase 2 (mitochondrial)
|
|
chr17_+_40688189
|
1.246
|
|
NAGLU
|
N-acetylglucosaminidase, alpha
|
|
chr6_+_43737937
|
1.244
|
NM_001025366
NM_001025367
NM_001025368
NM_001025369
NM_001025370
NM_001033756
NM_001171622
NM_001171623
NM_001171624
NM_001171625
NM_001171626
NM_001171627
NM_001171628
NM_001171629
NM_001171630
NM_001204384
NM_001204385
NM_003376
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr11_-_18258383
|
1.236
|
NM_006512
|
SAA4
|
serum amyloid A4, constitutive
|
|
chr6_-_155635611
|
1.227
|
NM_016020
|
TFB1M
|
transcription factor B1, mitochondrial
|
|
chr5_-_54830369
|
1.200
|
NM_003711
NM_176895
|
RNF138P1
PPAP2A
|
ring finger protein 138 pseudogene 1
phosphatidic acid phosphatase type 2A
|
|
chr6_+_42883726
|
1.179
|
NM_001243168
NM_001243169
NM_001243170
NM_138296
|
PTCRA
|
pre T-cell antigen receptor alpha
|
|
chr17_+_71161110
|
1.135
|
NM_001050
|
SSTR2
|
somatostatin receptor 2
|
|
chr11_+_71903172
|
1.122
|
NM_016729
|
FOLR1
|
folate receptor 1 (adult)
|
|
chr6_+_31913708
|
1.112
|
NM_001710
|
CFB
|
complement factor B
|
|
chr9_+_120466453
|
1.101
|
NM_138554
|
TLR4
|
toll-like receptor 4
|
|
chr7_-_126883568
|
1.094
|
NM_000845
|
GRM8
|
glutamate receptor, metabotropic 8
|
|
chr6_-_32634287
|
1.069
|
NM_001243961
NM_002123
|
HLA-DQB1
|
major histocompatibility complex, class II, DQ beta 1
|
|
chrX_-_2882288
|
1.064
|
NM_000047
|
ARSE
|
arylsulfatase E (chondrodysplasia punctata 1)
|
|
chr14_+_94492747
|
1.054
|
|
OTUB2
|
OTU domain, ubiquitin aldehyde binding 2
|
|
chr14_+_74815194
|
1.053
|
|
VRTN
|
vertebrae development homolog (pig)
|
|
chr12_-_53600999
|
1.045
|
NM_000889
|
ITGB7
|
integrin, beta 7
|
|
chr19_+_40093163
|
1.024
|
NM_013268
|
LGALS13
|
lectin, galactoside-binding, soluble, 13
|
|
chr19_-_10491172
|
1.001
|
|
TYK2
|
tyrosine kinase 2
|
|
chr11_-_119599253
|
0.968
|
NM_002855
NM_203285
NM_203286
|
PVRL1
|
poliovirus receptor-related 1 (herpesvirus entry mediator C)
|
|
chr8_-_82395401
|
0.963
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr15_+_40509628
|
0.958
|
NM_001128628
NM_001128629
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6
|
|
chr13_+_102104941
|
0.937
|
NM_004791
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains)
|
|
chr10_+_102295700
|
0.923
|
|
HIF1AN
|
hypoxia inducible factor 1, alpha subunit inhibitor
|
|
chr2_-_191872322
|
0.909
|
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa
|
|
chr17_-_3499124
|
0.906
|
NM_080705
|
TRPV1
|
transient receptor potential cation channel, subfamily V, member 1
|
|
chr22_-_33402648
|
0.863
|
NM_003490
NM_133633
|
SYN3
|
synapsin III
|
|
chr10_+_102729249
|
0.842
|
|
SEMA4G
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G
|
|
chr6_+_31913920
|
0.808
|
|
CFB
|
complement factor B
|
|
chr6_+_43738177
|
0.745
|
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr15_-_78369825
|
0.726
|
NM_015079
NM_144572
|
TBC1D2B
|
TBC1 domain family, member 2B
|
|
chr3_+_132318980
|
0.720
|
NM_178445
|
CCRL1
|
chemokine (C-C motif) receptor-like 1
|
|
chr11_-_4629371
|
0.700
|
NM_018073
|
TRIM68
|
tripartite motif containing 68
|
|
chr4_+_39408472
|
0.688
|
NM_175737
|
KLB
|
klotho beta
|
|
chr2_-_37193609
|
0.654
|
NM_003162
|
STRN
|
striatin, calmodulin binding protein
|
|
chr17_+_32646058
|
0.619
|
NM_005623
|
CCL8
|
chemokine (C-C motif) ligand 8
|
|
chr6_+_121756744
|
0.609
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr12_-_10588543
|
0.605
|
NM_002260
|
KLRC2
KLRC3
|
killer cell lectin-like receptor subfamily C, member 2
killer cell lectin-like receptor subfamily C, member 3
|
|
chr12_-_57351381
|
0.605
|
NM_003708
|
RDH16
|
retinol dehydrogenase 16 (all-trans)
|
|
chr6_-_30128688
|
0.583
|
NM_006778
NM_052828
|
TRIM10
|
tripartite motif containing 10
|
|
chr6_+_43738394
|
0.564
|
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr2_+_102927961
|
0.540
|
NM_016232
|
IL18R1
IL1RL1
|
interleukin 18 receptor 1
interleukin 1 receptor-like 1
|
|
chr6_-_110736748
|
0.539
|
NM_003649
NM_004032
|
DDO
|
D-aspartate oxidase
|
|
chr8_+_28174648
|
0.511
|
NM_006228
|
PNOC
|
prepronociceptin
|
|
chr17_+_26989193
|
0.506
|
|
SUPT6H
|
suppressor of Ty 6 homolog (S. cerevisiae)
|
|
chr3_-_184095927
|
0.490
|
NM_000460
NM_001177597
NM_001177598
|
THPO
|
thrombopoietin
|
|
chr10_+_101542554
|
0.487
|
|
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2
|
|
chrX_+_12924738
|
0.452
|
NM_138636
|
TLR8
|
toll-like receptor 8
|
|
chrX_+_119495939
|
0.445
|
NM_001142447
NM_012069
|
ATP1B4
|
ATPase, Na+/K+ transporting, beta 4 polypeptide
|
|
chr1_+_46640748
|
0.439
|
NM_005727
|
TSPAN1
|
tetraspanin 1
|
|
chr22_+_21771661
|
0.433
|
NM_015094
|
HIC2
|
hypermethylated in cancer 2
|
|
chr2_+_233243243
|
0.413
|
NM_001632
|
ALPP
|
alkaline phosphatase, placental
|
|
chr12_-_56848398
|
0.410
|
NM_012064
|
MIP
|
major intrinsic protein of lens fiber
|
|
chr15_-_66084438
|
0.403
|
|
DENND4A
|
DENN/MADD domain containing 4A
|
|
chr19_+_55417507
|
0.374
|
NM_001145457
NM_001145458
NM_001242356
NM_001242357
NM_004829
|
NCR1
|
natural cytotoxicity triggering receptor 1
|
|
chr1_-_150602089
|
0.364
|
NM_004436
NM_207042
NM_207043
NM_207044
NM_207168
|
ENSA
|
endosulfine alpha
|
|
chr3_-_17741511
|
0.321
|
NM_001134381
|
TBC1D5
|
TBC1 domain family, member 5
|
|
chr8_+_17396285
|
0.317
|
NM_001164771
NM_003046
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr11_+_5710816
|
0.218
|
NM_001199573
NM_006074
|
TRIM22
|
tripartite motif containing 22
|
|
chr5_+_64859098
|
0.207
|
NM_015342
|
PPWD1
|
peptidylprolyl isomerase domain and WD repeat containing 1
|
|
chr1_-_19217384
|
0.189
|
NM_001161504
|
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1
|
|
chr19_-_51220194
|
0.176
|
NM_016148
|
SHANK1
|
SH3 and multiple ankyrin repeat domains 1
|
|
chr1_+_178310605
|
0.168
|
NM_004841
|
RASAL2
|
RAS protein activator like 2
|
|
chr6_-_35696359
|
0.156
|
NM_001145775
|
FKBP5
|
FK506 binding protein 5
|
|
chrX_-_24690740
|
0.151
|
NM_001163264
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr7_-_107880492
|
0.135
|
NM_001037132
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr2_-_216003068
|
0.133
|
NM_173076
|
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12
|