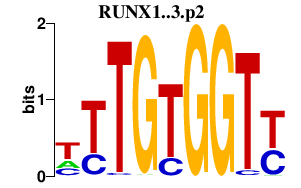
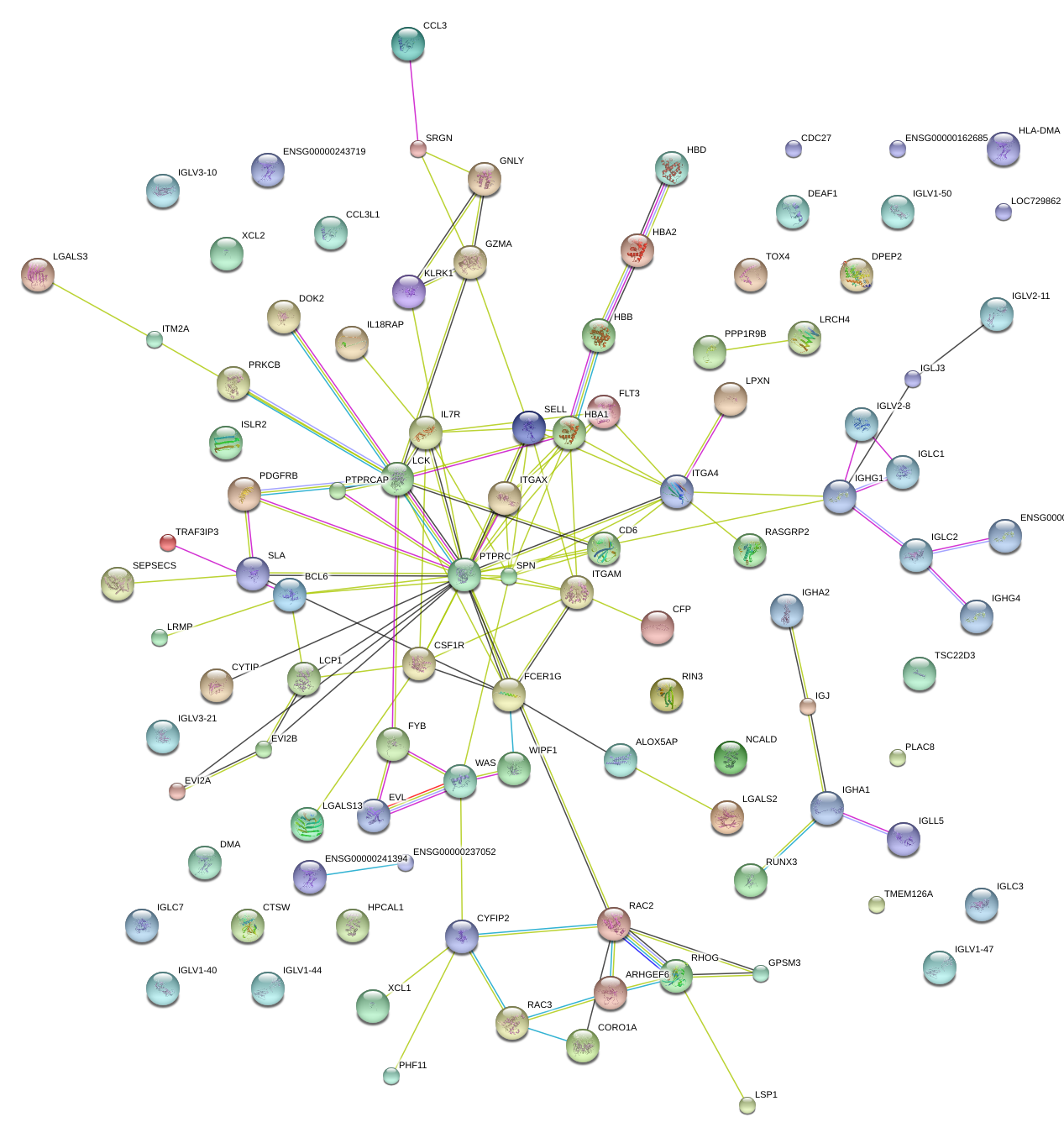
|
chr17_-_29641068
|
78.814
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr17_-_29641094
|
77.284
|
NM_006495
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr17_-_29641087
|
72.716
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr11_+_1889902
|
66.081
|
NM_001013253
|
LSP1
|
lymphocyte-specific protein 1
|
|
chr16_+_23847321
|
56.737
|
|
PRKCB
|
protein kinase C, beta
|
|
chr1_-_25291474
|
56.307
|
NM_001031680
|
RUNX3
|
runt-related transcription factor 3
|
|
chr6_-_32163299
|
54.095
|
NM_022107
|
GPSM3
|
G-protein signaling modulator 3
|
|
chr1_+_198608136
|
52.962
|
NM_002838
NM_080921
NM_080923
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr1_+_198608216
|
51.668
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr16_+_23847271
|
51.537
|
NM_002738
NM_212535
|
PRKCB
|
protein kinase C, beta
|
|
chr1_+_198608244
|
49.515
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr16_+_30194925
|
48.242
|
|
CORO1A
|
coronin, actin binding protein, 1A
|
|
chrX_-_106960028
|
48.165
|
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr10_+_70847857
|
47.688
|
|
SRGN
|
serglycin
|
|
chr2_-_158300517
|
46.936
|
NM_004288
|
CYTIP
|
cytohesin 1 interacting protein
|
|
chr4_-_84035908
|
46.789
|
NM_001130715
NM_016619
|
PLAC8
|
placenta-specific 8
|
|
chr4_-_84035888
|
46.567
|
|
PLAC8
|
placenta-specific 8
|
|
chr11_-_58345593
|
44.450
|
NM_001143995
|
LPXN
|
leupaxin
|
|
chr10_+_70847817
|
42.851
|
NM_002727
|
SRGN
|
serglycin
|
|
chr1_+_209929378
|
41.935
|
NM_025228
|
TRAF3IP3
|
TRAF3 interacting protein 3
|
|
chr8_-_134072404
|
40.408
|
NM_006748
|
SLA
|
Src-like-adaptor
|
|
chrX_+_48542159
|
39.503
|
NM_000377
|
WAS
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia)
|
|
chr12_-_10542616
|
39.389
|
NM_007360
|
KLRK1
|
killer cell lectin-like receptor subfamily K, member 1
|
|
chr11_-_58343308
|
38.127
|
NM_004811
|
LPXN
|
leupaxin
|
|
chr16_-_68033355
|
37.108
|
NM_022355
|
DPEP2
|
dipeptidase 2
|
|
chrX_-_106960268
|
36.768
|
NM_004089
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr22_+_23222897
|
36.590
|
|
CYAT1
|
immunoglobulin lambda light chain-like
|
|
chr22_-_37640287
|
34.723
|
NM_002872
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr16_+_23847359
|
33.684
|
|
PRKCB
|
protein kinase C, beta
|
|
chr13_-_46756295
|
33.013
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr11_-_67205151
|
32.707
|
NM_005608
|
PTPRCAP
|
protein tyrosine phosphatase, receptor type, C-associated protein
|
|
chr17_-_34524054
|
32.559
|
NM_001001437
NM_021006
|
CCL3L3
CCL3
CCL3L1
|
chemokine (C-C motif) ligand 3-like 3
chemokine (C-C motif) ligand 3
chemokine (C-C motif) ligand 3-like 1
|
|
chr1_+_32716839
|
31.444
|
NM_005356
|
LCK
|
lymphocyte-specific protein tyrosine kinase
|
|
chr11_+_65647283
|
31.276
|
NM_001335
|
CTSW
|
cathepsin W
|
|
chr22_+_22786283
|
30.942
|
|
|
|
|
chr2_-_89165652
|
30.354
|
|
|
|
|
chr1_-_168513201
|
29.464
|
NM_003175
|
XCL1
XCL2
|
chemokine (C motif) ligand 1
chemokine (C motif) ligand 2
|
|
chrX_-_135863104
|
27.733
|
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr5_+_35856990
|
27.640
|
NM_002185
|
IL7R
|
interleukin 7 receptor
|
|
chrX_-_135863033
|
27.199
|
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr17_-_29648631
|
26.936
|
NM_001003927
NM_014210
|
EVI2A
|
ecotropic viral integration site 2A
|
|
chr2_+_85921413
|
26.896
|
NM_006433
NM_012483
|
GNLY
|
granulysin
|
|
chr22_+_22712091
|
26.854
|
|
IGLV1-44
IGL@
|
immunoglobulin lambda variable 1-44
immunoglobulin lambda locus
|
|
chr16_+_226678
|
26.794
|
NM_000558
|
HBA1
HBA2
|
hemoglobin, alpha 1
hemoglobin, alpha 2
|
|
chr22_-_37640187
|
25.456
|
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr16_+_29674564
|
25.334
|
|
SPN
|
sialophorin
|
|
chrX_-_135863502
|
25.276
|
NM_004840
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr1_+_161185084
|
24.493
|
NM_004106
|
FCER1G
|
Fc fragment of IgE, high affinity I, receptor for; gamma polypeptide
|
|
chr2_-_175462428
|
24.477
|
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr11_+_60739114
|
24.097
|
NM_006725
|
CD6
|
CD6 molecule
|
|
chr4_-_71532206
|
23.872
|
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides
|
|
chr14_+_100531764
|
23.628
|
|
EVL
|
Enah/Vasp-like
|
|
chrX_-_78623006
|
23.316
|
NM_001171581
NM_004867
|
ITM2A
|
integral membrane protein 2A
|
|
chr22_+_23054862
|
23.215
|
|
IGLV3-21
IGLJ3
IGLC1
|
immunoglobulin lambda variable 3-21
immunoglobulin lambda joining 3
immunoglobulin lambda constant 1 (Mcg marker)
|
|
chr13_+_50070527
|
23.139
|
NM_001040444
|
PHF11
|
PHD finger protein 11
|
|
chr8_-_21771111
|
23.089
|
NM_003974
|
DOK2
|
docking protein 2, 56kDa
|
|
chr1_+_161185061
|
22.926
|
|
FCER1G
|
Fc fragment of IgE, high affinity I, receptor for; gamma polypeptide
|
|
chr7_-_100183753
|
22.793
|
NM_002319
|
LRCH4
|
leucine-rich repeats and calponin homology (CH) domain containing 4
|
|
chr5_-_39203061
|
22.456
|
|
FYB
|
FYN binding protein
|
|
chr3_-_187454247
|
22.247
|
NM_001130845
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr4_-_71532341
|
22.211
|
NM_144646
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides
|
|
chr5_+_156696351
|
22.194
|
NM_014376
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr14_+_93118576
|
21.991
|
|
RIN3
|
Ras and Rab interactor 3
|
|
chr5_-_149466126
|
21.815
|
|
CSF1R
|
colony stimulating factor 1 receptor
|
|
chr5_-_149466094
|
21.396
|
|
CSF1R
|
colony stimulating factor 1 receptor
|
|
chr2_+_103035148
|
21.382
|
NM_003853
|
IL18RAP
|
interleukin 18 receptor accessory protein
|
|
chr5_-_149466173
|
21.265
|
|
CSF1R
|
colony stimulating factor 1 receptor
|
|
chr14_+_93118793
|
21.185
|
|
RIN3
|
Ras and Rab interactor 3
|
|
chr1_-_169680677
|
21.069
|
NM_000655
|
SELL
|
selectin L
|
|
chr14_+_100531745
|
21.044
|
NM_016337
|
EVL
|
Enah/Vasp-like
|
|
chr8_-_102803438
|
20.656
|
NM_032041
|
NCALD
|
neurocalcin delta
|
|
chr17_-_34417386
|
20.646
|
NM_002983
|
CCL3
|
chemokine (C-C motif) ligand 3
|
|
chr12_+_25205180
|
20.487
|
NM_001204126
NM_001204127
NM_006152
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr8_-_102803178
|
20.207
|
|
NCALD
|
neurocalcin delta
|
|
chr6_-_32920812
|
20.159
|
NM_006120
|
HLA-DMA
|
major histocompatibility complex, class II, DM alpha
|
|
chr5_-_39219578
|
19.740
|
NM_001465
NM_199335
|
FYB
|
FYN binding protein
|
|
chr16_+_31366468
|
19.710
|
NM_000887
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit)
|
|
chr19_-_36233311
|
19.667
|
NM_024660
|
IGFLR1
|
IGF-like family receptor 1
|
|
chr15_+_74422589
|
19.480
|
NM_001130138
|
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2
|
|
chr22_-_37975937
|
19.438
|
NM_006498
|
LGALS2
|
lectin, galactoside-binding, soluble, 2
|
|
chrX_-_47489703
|
19.387
|
NM_002621
|
CFP
|
complement factor properdin
|
|
chr13_+_31309644
|
19.199
|
NM_001629
|
ALOX5AP
|
arachidonate 5-lipoxygenase-activating protein
|
|
chr2_+_182321618
|
18.463
|
NM_000885
|
ITGA4
|
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor)
|
|
chr8_-_102803196
|
18.047
|
|
NCALD
|
neurocalcin delta
|
|
chr19_-_18391440
|
17.860
|
|
|
|
|
chr16_+_28505920
|
17.658
|
NM_018690
|
APOBR
|
apolipoprotein B receptor
|
|
chr13_-_28674646
|
17.652
|
NM_004119
|
FLT3
|
fms-related tyrosine kinase 3
|
|
chr16_+_31271327
|
17.634
|
|
ITGAM
|
integrin, alpha M (complement component 3 receptor 3 subunit)
|
|
chr14_-_107013474
|
17.509
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr13_-_46756291
|
17.391
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr12_+_25205488
|
17.378
|
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr11_-_5248180
|
17.183
|
NM_000518
|
HBB
|
hemoglobin, beta
|
|
chr11_-_64512925
|
17.126
|
NM_153819
|
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated)
|
|
chr5_-_39270708
|
17.094
|
NM_001243093
|
FYB
|
FYN binding protein
|
|
chr5_+_54398473
|
16.843
|
NM_006144
|
GZMA
|
granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3)
|
|
chr16_+_31271311
|
16.721
|
|
ITGAM
|
integrin, alpha M (complement component 3 receptor 3 subunit)
|
|
chrX_+_48542193
|
16.699
|
|
WAS
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia)
|
|
chr5_-_149492934
|
16.290
|
NM_005211
|
CSF1R
|
colony stimulating factor 1 receptor
|
|
chr17_+_72462521
|
16.167
|
NM_007261
|
CD300A
|
CD300a molecule
|
|
chr10_+_11047274
|
16.157
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr12_-_51717941
|
15.744
|
|
BIN2
|
bridging integrator 2
|
|
chr12_-_54121203
|
15.399
|
NM_001143682
NM_020898
|
CALCOCO1
|
calcium binding and coiled-coil domain 1
|
|
chr13_-_41593491
|
15.369
|
NM_172373
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr22_+_37257004
|
15.343
|
NM_000631
NM_013416
|
NCF4
|
neutrophil cytosolic factor 4, 40kDa
|
|
chr12_-_51717905
|
15.286
|
NM_016293
|
BIN2
|
bridging integrator 2
|
|
chr1_-_150738255
|
15.119
|
|
CTSS
|
cathepsin S
|
|
chr12_-_112819755
|
15.046
|
NM_001109662
|
C12orf51
|
chromosome 12 open reading frame 51
|
|
chr2_-_89442551
|
14.732
|
|
IGKC
IGK@
|
immunoglobulin kappa constant
immunoglobulin kappa locus
|
|
chr10_+_11047253
|
14.712
|
NM_001025077
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr1_+_174843523
|
14.711
|
NM_001243763
|
RABGAP1L
|
RAB GTPase activating protein 1-like
|
|
chr17_-_1532109
|
14.680
|
NM_152346
|
SLC43A2
|
solute carrier family 43, member 2
|
|
chr22_+_23154240
|
14.608
|
|
|
|
|
chr5_-_137071462
|
14.590
|
|
KLHL3
|
kelch-like 3 (Drosophila)
|
|
chr13_-_41593405
|
14.547
|
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr15_-_22473092
|
14.457
|
|
|
|
|
chr5_-_176326314
|
14.454
|
NM_002115
|
HK3
|
hexokinase 3 (white cell)
|
|
chr10_-_126432893
|
14.317
|
NM_014661
|
FAM53B
|
family with sequence similarity 53, member B
|
|
chr19_-_2151332
|
14.236
|
|
AP3D1
|
adaptor-related protein complex 3, delta 1 subunit
|
|
chr1_+_32739711
|
14.053
|
NM_001042771
|
LCK
|
lymphocyte-specific protein tyrosine kinase
|
|
chr14_-_106780535
|
13.959
|
|
IGHA1
IGHA2
IGHD
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr9_-_123688352
|
13.877
|
|
TRAF1
|
TNF receptor-associated factor 1
|
|
chr21_+_42733916
|
13.798
|
NM_002463
|
LOC100131190
MX2
|
uncharacterized LOC100131190
myxovirus (influenza virus) resistance 2 (mouse)
|
|
chr2_+_68592432
|
13.565
|
|
PLEK
|
pleckstrin
|
|
chr19_+_55085273
|
13.552
|
|
LILRA2
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 2
|
|
chr2_+_68592317
|
13.461
|
NM_002664
|
PLEK
|
pleckstrin
|
|
chr19_+_51728369
|
13.355
|
|
CD33
|
CD33 molecule
|
|
chr1_-_92951593
|
13.318
|
NM_001127216
|
GFI1
|
growth factor independent 1 transcription repressor
|
|
chr13_+_31309690
|
13.239
|
|
ALOX5AP
|
arachidonate 5-lipoxygenase-activating protein
|
|
chr19_-_3786221
|
13.017
|
|
MATK
|
megakaryocyte-associated tyrosine kinase
|
|
chr19_+_50015535
|
12.981
|
NM_001136019
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr19_+_47813074
|
12.953
|
NM_001736
|
C5AR1
|
complement component 5a receptor 1
|
|
chr1_-_203155827
|
12.925
|
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39)
|
|
chr19_-_54746599
|
12.901
|
NM_024318
|
LILRB3
LILRA6
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 6
|
|
chr16_+_31271277
|
12.866
|
NM_000632
NM_001145808
|
ITGAM
|
integrin, alpha M (complement component 3 receptor 3 subunit)
|
|
chr1_+_168545710
|
12.854
|
NM_002995
|
XCL1
|
chemokine (C motif) ligand 1
|
|
chr13_-_46756326
|
12.687
|
NM_002298
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr1_+_192544856
|
12.623
|
NM_002922
|
RGS1
|
regulator of G-protein signaling 1
|
|
chr4_+_8951476
|
12.601
|
NM_001040071
|
LOC650293
|
seven transmembrane helix receptor
|
|
chr2_+_90077732
|
12.572
|
|
|
|
|
chr11_+_2323236
|
12.551
|
NM_139022
|
TSPAN32
|
tetraspanin 32
|
|
chr11_-_128457452
|
12.537
|
NM_001143820
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr17_-_1531543
|
12.477
|
|
SLC43A2
|
solute carrier family 43, member 2
|
|
chr1_-_203155867
|
12.453
|
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39)
|
|
chr4_-_38806410
|
12.382
|
NM_003263
|
TLR1
|
toll-like receptor 1
|
|
chr5_+_75699003
|
12.378
|
NM_006633
|
IQGAP2
|
IQ motif containing GTPase activating protein 2
|
|
chr11_-_5275904
|
12.351
|
NM_000184
|
HBG1
HBG2
|
hemoglobin, gamma A
hemoglobin, gamma G
|
|
chr16_-_28621279
|
12.321
|
NM_001055
NM_177529
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1
|
|
chr6_-_35696359
|
12.112
|
NM_001145775
|
FKBP5
|
FK506 binding protein 5
|
|
chr17_+_45810609
|
12.062
|
NM_013351
|
TBX21
|
T-box 21
|
|
chr8_+_1952875
|
12.030
|
|
KBTBD11
|
kelch repeat and BTB (POZ) domain containing 11
|
|
chr20_+_44637539
|
11.982
|
NM_004994
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase)
|
|
chr2_+_128175989
|
11.807
|
NM_000312
|
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa)
|
|
chr19_-_18392048
|
11.787
|
|
JUND
|
jun D proto-oncogene
|
|
chr11_+_67033929
|
11.735
|
|
ADRBK1
|
adrenergic, beta, receptor kinase 1
|
|
chr19_+_50015868
|
11.725
|
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr3_-_150920946
|
11.631
|
NM_013308
|
GPR171
|
G protein-coupled receptor 171
|
|
chr6_-_31550065
|
11.608
|
NM_002341
NM_009588
|
LTB
|
lymphotoxin beta (TNF superfamily, member 3)
|
|
chr6_+_31982538
|
11.526
|
NM_001242823
NM_001252204
NM_007293
NM_001002029
|
LOC100293534
C4A
C4B
|
complement C4-B-like
complement component 4A (Rodgers blood group)
complement component 4B (Chido blood group)
|
|
chr3_-_151047309
|
11.525
|
NM_176894
|
P2RY13
|
purinergic receptor P2Y, G-protein coupled, 13
|
|
chr14_+_74551655
|
11.456
|
NM_001024674
|
LIN52
|
lin-52 homolog (C. elegans)
|
|
chr21_-_36421630
|
11.395
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr19_+_3136026
|
11.376
|
NM_002068
|
GNA15
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class)
|
|
chr4_-_74847669
|
11.159
|
NM_002619
|
PF4
|
platelet factor 4
|
|
chr1_-_160832466
|
11.152
|
NM_001166663
NM_001166664
NM_016382
|
CD244
|
CD244 molecule, natural killer cell receptor 2B4
|
|
chr15_-_34875060
|
11.036
|
|
GOLGA8B
|
golgin A8 family, member B
|
|
chr12_-_51717835
|
11.034
|
|
BIN2
|
bridging integrator 2
|
|
chr2_+_182322265
|
11.033
|
|
ITGA4
|
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor)
|
|
chr22_-_19419177
|
11.012
|
|
HIRA
|
HIR histone cell cycle regulation defective homolog A (S. cerevisiae)
|
|
chr1_+_17634689
|
10.948
|
NM_012387
|
PADI4
|
peptidyl arginine deiminase, type IV
|
|
chr19_-_3786382
|
10.812
|
NM_139354
NM_139355
|
MATK
|
megakaryocyte-associated tyrosine kinase
|
|
chr22_-_31688329
|
10.806
|
NM_001135911
NM_052880
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1
|
|
chr11_-_5255712
|
10.742
|
NM_000519
|
HBD
|
hemoglobin, delta
|
|
chr12_-_109025825
|
10.729
|
NM_001206609
|
SELPLG
|
selectin P ligand
|
|
chr22_+_40297183
|
10.686
|
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr1_-_203155910
|
10.650
|
NM_001276
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39)
|
|
chr15_+_75494235
|
10.633
|
|
C15orf39
|
chromosome 15 open reading frame 39
|
|
chr11_+_46366816
|
10.616
|
NM_201533
|
DGKZ
|
diacylglycerol kinase, zeta
|
|
chr1_-_173174611
|
10.614
|
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4
|
|
chr15_-_40600025
|
10.313
|
|
PLCB2
|
phospholipase C, beta 2
|
|
chr17_-_56606710
|
10.269
|
NM_004574
|
SEPT4
|
septin 4
|
|
chr6_+_31949800
|
10.240
|
NM_001242823
NM_001252204
NM_007293
NM_001002029
|
LOC100293534
C4A
C4B
|
complement C4-B-like
complement component 4A (Rodgers blood group)
complement component 4B (Chido blood group)
|
|
chr6_+_72926462
|
10.230
|
NM_001168410
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr1_-_203155745
|
10.222
|
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39)
|
|
chr19_-_2151551
|
10.220
|
NM_001077523
NM_003938
|
AP3D1
|
adaptor-related protein complex 3, delta 1 subunit
|
|
chr18_+_77253324
|
10.107
|
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr19_+_51645556
|
10.056
|
NM_014385
NM_016543
|
SIGLEC7
|
sialic acid binding Ig-like lectin 7
|
|
chr2_-_74756823
|
10.041
|
|
AUP1
|
ancient ubiquitous protein 1
|
|
chr22_+_40297085
|
10.019
|
NM_004810
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr10_+_129845806
|
10.011
|
NM_130435
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E
|
|
chr1_-_207095198
|
10.006
|
NM_001193338
NM_001142473
NM_005449
|
FAIM3
|
Fas apoptotic inhibitory molecule 3
|
|
chr14_+_21423627
|
10.004
|
NM_002934
|
RNASE2
|
ribonuclease, RNase A family, 2 (liver, eosinophil-derived neurotoxin)
|
|
chr6_-_32784727
|
9.998
|
NM_002120
|
HLA-DOB
|
major histocompatibility complex, class II, DO beta
|
|
chr9_+_123963728
|
9.915
|
|
GSN
|
gelsolin
|
|
chr22_-_19418998
|
9.890
|
|
HIRA
|
HIR histone cell cycle regulation defective homolog A (S. cerevisiae)
|
|
chr22_-_19419215
|
9.756
|
NM_003325
|
HIRA
|
HIR histone cell cycle regulation defective homolog A (S. cerevisiae)
|
|
chr15_+_75494233
|
9.713
|
|
C15orf39
|
chromosome 15 open reading frame 39
|
|
chr19_+_51728319
|
9.601
|
NM_001082618
NM_001177608
NM_001772
|
CD33
|
CD33 molecule
|
|
chr17_-_79008372
|
9.578
|
|
FLJ90757
|
uncharacterized LOC440465
|
|
chr11_+_67033889
|
9.544
|
NM_001619
|
ADRBK1
|
adrenergic, beta, receptor kinase 1
|
|
chr22_+_35695267
|
9.484
|
NM_001135729
|
TOM1
|
target of myb1 (chicken)
|