|
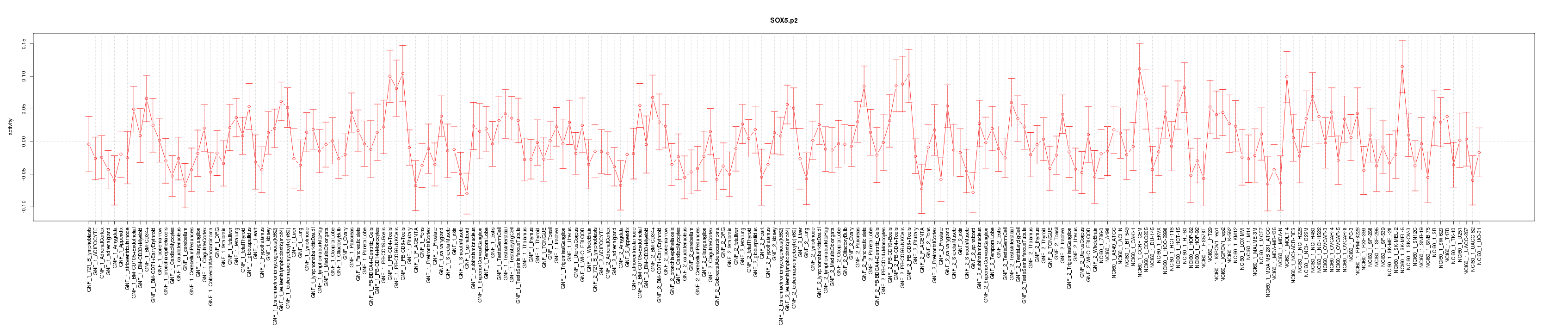
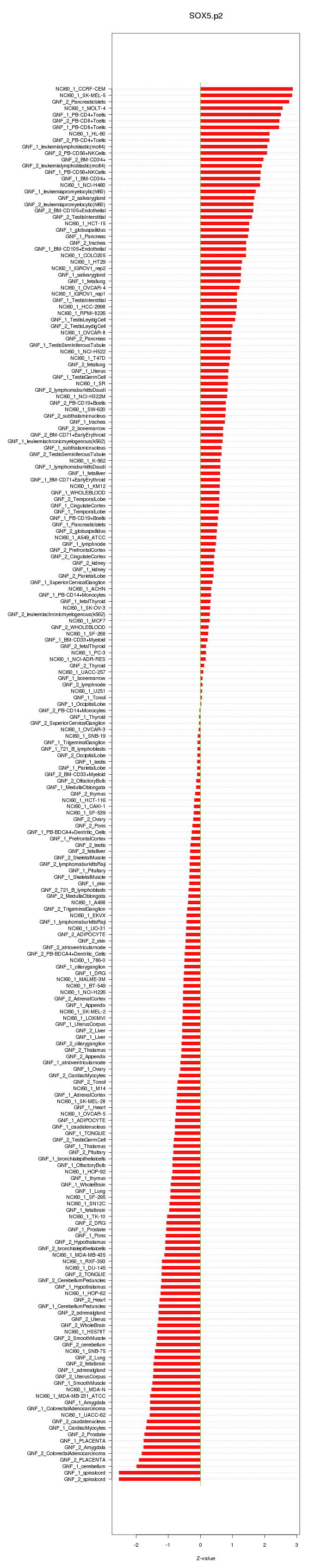
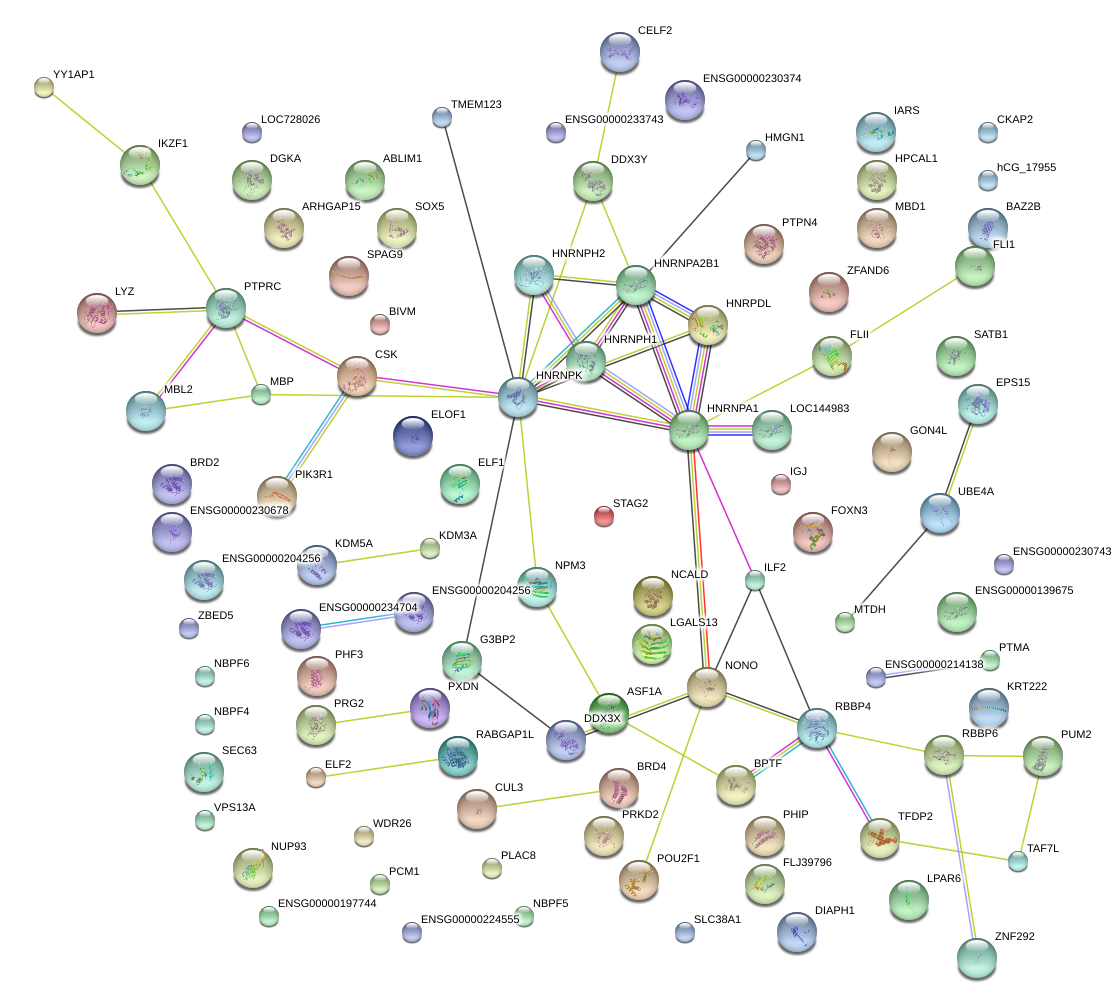
chr1_+_198608244
|
34.572
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr1_+_198608136
|
29.166
|
NM_002838
NM_080921
NM_080923
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr1_+_198608216
|
24.609
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chrX_+_123234462
|
23.361
|
|
STAG2
|
stromal antigen 2
|
|
chr9_-_86584079
|
20.529
|
|
HNRNPK
|
heterogeneous nuclear ribonucleoprotein K
|
|
chr16_+_56817367
|
17.165
|
NM_001242796
|
NUP93
|
nucleoporin 93kDa
|
|
chr12_+_69742130
|
16.381
|
NM_000239
|
LYZ
|
lysozyme
|
|
chr4_-_83347222
|
15.759
|
|
HNRPDL
|
heterogeneous nuclear ribonucleoprotein D-like
|
|
chr17_+_65822242
|
15.576
|
|
BPTF
|
bromodomain PHD finger transcription factor
|
|
chr10_+_11206928
|
15.172
|
NM_001025076
NM_001083591
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr17_+_65822206
|
14.893
|
|
BPTF
|
bromodomain PHD finger transcription factor
|
|
chr12_+_69742140
|
14.095
|
|
LYZ
|
lysozyme
|
|
chr2_+_143886898
|
12.800
|
NM_018460
|
ARHGAP15
|
Rho GTPase activating protein 15
|
|
chr7_-_26232148
|
12.687
|
|
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1
|
|
chr16_+_56815703
|
12.261
|
NM_001242795
|
NUP93
|
nucleoporin 93kDa
|
|
chr2_+_143886969
|
12.087
|
|
ARHGAP15
|
Rho GTPase activating protein 15
|
|
chr2_-_225370687
|
12.063
|
|
CUL3
|
cullin 3
|
|
chrY_+_15016018
|
11.132
|
NM_001122665
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked
|
|
chr10_+_11207053
|
9.640
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr1_+_174844655
|
9.572
|
NM_001035230
|
RABGAP1L
|
RAB GTPase activating protein 1-like
|
|
chrX_+_70503041
|
9.512
|
NM_001145408
NM_001145409
NM_001145410
NM_007363
|
NONO
|
non-POU domain containing, octamer-binding
|
|
chr8_+_17822093
|
9.058
|
|
PCM1
|
pericentriolar material 1
|
|
chr13_+_103459495
|
8.824
|
NM_001204425
|
BIVM-ERCC5
BIVM
|
BIVM-ERCC5 readthrough
basic, immunoglobulin-like variable motif containing
|
|
chr18_-_74728677
|
8.704
|
|
MBP
|
myelin basic protein
|
|
chr11_+_118257222
|
8.680
|
|
UBE4A
|
ubiquitination factor E4A
|
|
chr17_-_38821286
|
8.408
|
NM_152349
|
KRT222
|
keratin 222
|
|
chr6_+_119215240
|
8.356
|
NM_014034
|
ASF1A
|
ASF1 anti-silencing function 1 homolog A (S. cerevisiae)
|
|
chr2_+_86668488
|
8.051
|
NM_001146688
|
KDM3A
|
lysine (K)-specific demethylase 3A
|
|
chr2_-_20512016
|
7.851
|
|
PUM2
|
pumilio homolog 2 (Drosophila)
|
|
chr5_-_179044835
|
7.715
|
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H)
|
|
chr15_+_80364902
|
7.638
|
NM_001242915
NM_001242916
|
ZFAND6
|
zinc finger, AN1-type domain 6
|
|
chr9_+_79968344
|
7.619
|
|
VPS13A
|
vacuolar protein sorting 13 homolog A (S. cerevisiae)
|
|
chr1_-_224622000
|
7.613
|
NM_001115113
NM_025160
|
WDR26
|
WD repeat domain 26
|
|
chr13_+_53035227
|
7.607
|
|
CKAP2
|
cytoskeleton associated protein 2
|
|
chr3_-_141747434
|
7.582
|
NM_001178141
NM_001178142
NM_006286
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr2_+_86668428
|
7.480
|
|
KDM3A
|
lysine (K)-specific demethylase 3A
|
|
chr2_-_74382352
|
7.264
|
|
|
|
|
chr3_-_18480203
|
7.098
|
NM_001131010
|
SATB1
|
SATB homeobox 1
|
|
chr17_-_49124128
|
7.095
|
NM_001251971
|
SPAG9
|
sperm associated antigen 9
|
|
chr1_-_51887740
|
7.091
|
NM_001159969
|
EPS15
|
epidermal growth factor receptor pathway substrate 15
|
|
chr6_+_87969619
|
7.075
|
|
ZNF292
|
zinc finger protein 292
|
|
chr6_-_79675672
|
7.066
|
|
PHIP
|
pleckstrin homology domain interacting protein
|
|
chr13_-_41593405
|
7.049
|
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr5_-_140895407
|
7.011
|
|
DIAPH1
|
diaphanous homolog 1 (Drosophila)
|
|
chr13_-_48987652
|
6.973
|
NM_001162498
|
LPAR6
|
lysophosphatidic acid receptor 6
|
|
chr1_+_33116748
|
6.959
|
NM_001135255
NM_005610
NM_001135256
|
RBBP4
|
retinoblastoma binding protein 4
|
|
chr11_-_102323353
|
6.865
|
NM_052932
|
TMEM123
|
transmembrane protein 123
|
|
chr2_-_20527120
|
6.851
|
NM_015317
|
PUM2
|
pumilio homolog 2 (Drosophila)
|
|
chr1_-_153643456
|
6.850
|
|
ILF2
|
interleukin enhancer binding factor 2, 45kDa
|
|
chr12_-_46633551
|
6.783
|
|
SLC38A1
|
solute carrier family 38, member 1
|
|
chr13_-_41593491
|
6.738
|
NM_172373
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr2_+_120517206
|
6.563
|
NM_002830
|
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte)
|
|
chr12_-_10562744
|
6.553
|
NM_001199805
|
KLRC4-KLRK1
|
KLRC4-KLRK1 readthrough
|
|
chr12_-_498256
|
6.435
|
|
KDM5A
|
lysine (K)-specific demethylase 5A
|
|
chr1_+_167298280
|
6.419
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr10_-_116444374
|
6.396
|
NM_001003407
NM_001003408
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr4_-_76598563
|
6.353
|
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2
|
|
chr8_-_102803178
|
6.338
|
|
NCALD
|
neurocalcin delta
|
|
chr15_+_75074714
|
6.307
|
|
CSK
|
c-src tyrosine kinase
|
|
chr4_-_140060601
|
6.304
|
NM_201999
|
ELF2
|
E74-like factor 2 (ets domain transcription factor)
|
|
chr1_-_153643474
|
6.295
|
NM_004515
|
ILF2
|
interleukin enhancer binding factor 2, 45kDa
|
|
chr1_-_153643433
|
6.220
|
|
ILF2
|
interleukin enhancer binding factor 2, 45kDa
|
|
chr2_+_86668270
|
6.176
|
NM_018433
|
KDM3A
|
lysine (K)-specific demethylase 3A
|
|
chr1_-_153643428
|
6.174
|
|
ILF2
|
interleukin enhancer binding factor 2, 45kDa
|
|
chr16_+_24551672
|
6.110
|
|
RBBP6
|
retinoblastoma binding protein 6
|
|
chr1_-_153643417
|
6.097
|
|
ILF2
|
interleukin enhancer binding factor 2, 45kDa
|
|
chr7_+_50344255
|
6.054
|
NM_001220765
NM_001220766
NM_001220767
NM_001220768
NM_001220769
NM_001220770
NM_001220771
NM_001220772
NM_001220773
NM_001220774
NM_001220775
NM_001220776
NM_006060
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr9_-_95055913
|
6.046
|
|
IARS
|
isoleucyl-tRNA synthetase
|
|
chr8_+_98656494
|
5.959
|
|
MTDH
|
metadherin
|
|
chr1_-_224621714
|
5.940
|
|
WDR26
|
WD repeat domain 26
|
|
chr4_-_76598558
|
5.900
|
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2
|
|
chr6_-_108279320
|
5.811
|
|
SEC63
|
SEC63 homolog (S. cerevisiae)
|
|
chr8_+_98656428
|
5.729
|
|
MTDH
|
metadherin
|
|
chr5_+_67588395
|
5.621
|
NM_001242466
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr6_-_108279307
|
5.601
|
|
SEC63
|
SEC63 homolog (S. cerevisiae)
|
|
chr13_+_53216564
|
5.599
|
|
HNRNPA1L2
|
heterogeneous nuclear ribonucleoprotein A1-like 2
|
|
chr11_-_10879365
|
5.539
|
NM_001143667
NM_021211
|
ZBED5
|
zinc finger, BED-type containing 5
|
|
chr8_-_102803196
|
5.488
|
|
NCALD
|
neurocalcin delta
|
|
chr4_-_71532206
|
5.454
|
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides
|
|
chr6_-_108279212
|
5.442
|
|
SEC63
|
SEC63 homolog (S. cerevisiae)
|
|
chr2_+_232573226
|
5.421
|
NM_001099285
NM_002823
|
PTMA
|
prothymosin, alpha
|
|
chr2_-_160472960
|
5.340
|
NM_013450
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B
|
|
chr4_-_84035908
|
5.314
|
NM_001130715
NM_016619
|
PLAC8
|
placenta-specific 8
|
|
chr3_-_56698073
|
5.285
|
NM_015224
|
FAM208A
|
family with sequence similarity 208, member A
|
|
chr1_-_155649597
|
5.272
|
NM_001198899
|
YY1AP1
|
YY1 associated protein 1
|
|
chr6_+_32940860
|
5.120
|
NM_001199456
|
BRD2
|
bromodomain containing 2
|
|
chr11_+_128563772
|
5.107
|
NM_001167681
NM_002017
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr19_-_47217576
|
5.093
|
NM_001079882
|
PRKD2
|
protein kinase D2
|
|
chr14_-_89883306
|
5.079
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr8_+_98656329
|
5.027
|
NM_178812
|
MTDH
|
metadherin
|
|
chr6_+_64356430
|
5.004
|
NM_015153
|
PHF3
|
PHD finger protein 3
|
|
chr15_+_75074417
|
4.957
|
NM_001127190
NM_004383
|
CSK
|
c-src tyrosine kinase
|
|
chr12_+_56324945
|
4.956
|
NM_001345
|
DGKA
|
diacylglycerol kinase, alpha 80kDa
|
|
chr12_-_24103953
|
4.827
|
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr4_-_84035888
|
4.827
|
|
PLAC8
|
placenta-specific 8
|
|
chr17_-_73774447
|
4.823
|
|
|
|
|
chr1_-_108786663
|
4.803
|
NM_001143989
|
NBPF4
NBPF6
|
neuroblastoma breakpoint family, member 4
neuroblastoma breakpoint family, member 6
|
|
chr21_-_40721010
|
4.724
|
NM_004965
|
HMGN1
|
high mobility group nucleosome binding domain 1
|
|
chr5_-_71803119
|
4.722
|
NM_152625
|
ZNF366
|
zinc finger protein 366
|
|
chr6_-_24877453
|
4.646
|
NM_015864
|
FAM65B
|
family with sequence similarity 65, member B
|
|
chr4_+_175205044
|
4.614
|
NM_001145314
|
CEP44
|
centrosomal protein 44kDa
|
|
chrX_+_107288199
|
4.590
|
NM_001170553
NM_182607
|
VSIG1
|
V-set and immunoglobulin domain containing 1
|
|
chr4_-_83350548
|
4.553
|
|
HNRPDL
|
heterogeneous nuclear ribonucleoprotein D-like
|
|
chr7_-_104909442
|
4.473
|
NM_182691
|
SRPK2
|
SRSF protein kinase 2
|
|
chrX_-_135749436
|
4.443
|
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr12_-_122985396
|
4.443
|
|
ZCCHC8
|
zinc finger, CCHC domain containing 8
|
|
chr9_-_15510899
|
4.440
|
NM_001128217
NM_033222
|
PSIP1
|
PC4 and SFRS1 interacting protein 1
|
|
chr3_+_111260925
|
4.416
|
NM_005816
NM_198196
|
CD96
|
CD96 molecule
|
|
chr5_-_179051586
|
4.390
|
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H)
|
|
chrX_+_123094377
|
4.353
|
NM_001042749
|
STAG2
|
stromal antigen 2
|
|
chr3_+_151986731
|
4.350
|
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr6_-_99849266
|
4.297
|
|
PNISR
|
PNN-interacting serine/arginine-rich protein
|
|
chr1_+_62902735
|
4.274
|
|
USP1
|
ubiquitin specific peptidase 1
|
|
chr3_+_111261039
|
4.269
|
|
CD96
|
CD96 molecule
|
|
chr11_+_36317724
|
4.267
|
NM_001160167
|
PRR5L
|
proline rich 5 like
|
|
chr4_+_170581212
|
4.262
|
NM_001243374
|
CLCN3
|
chloride channel 3
|
|
chr20_-_2641476
|
4.238
|
|
IDH3B
|
isocitrate dehydrogenase 3 (NAD+) beta
|
|
chr10_-_75193297
|
4.231
|
NM_001024593
|
ZMYND17
|
zinc finger, MYND-type containing 17
|
|
chr20_+_32150139
|
4.197
|
NM_001039709
NM_005093
|
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2
|
|
chr5_-_175964223
|
4.175
|
NM_014901
|
RNF44
|
ring finger protein 44
|
|
chr6_+_56954978
|
4.143
|
|
ZNF451
|
zinc finger protein 451
|
|
chr5_+_65372744
|
4.127
|
|
ERBB2IP
|
erbb2 interacting protein
|
|
chr3_+_142342265
|
4.086
|
NM_002670
|
PLS1
|
plastin 1
|
|
chr20_-_45985411
|
4.084
|
NM_012408
NM_183047
NM_183048
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr1_+_167298382
|
4.073
|
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr21_+_17102370
|
4.068
|
NM_013396
|
USP25
|
ubiquitin specific peptidase 25
|
|
chrX_-_151619669
|
3.973
|
NM_000808
|
GABRA3
|
gamma-aminobutyric acid (GABA) A receptor, alpha 3
|
|
chr12_-_9913415
|
3.950
|
NM_001781
|
CD69
|
CD69 molecule
|
|
chr14_+_52456322
|
3.936
|
|
C14orf166
|
chromosome 14 open reading frame 166
|
|
chr7_+_111846642
|
3.909
|
NM_021994
|
ZNF277
|
zinc finger protein 277
|
|
chr14_-_20801427
|
3.896
|
NM_021178
NM_182849
|
CCNB1IP1
|
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase
|
|
chr4_-_83350452
|
3.883
|
|
HNRPDL
|
heterogeneous nuclear ribonucleoprotein D-like
|
|
chr10_-_103874661
|
3.862
|
NM_003893
|
LDB1
|
LIM domain binding 1
|
|
chr6_+_37137882
|
3.860
|
NM_001243186
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr7_-_16844606
|
3.825
|
NM_006408
|
AGR2
|
anterior gradient 2 homolog (Xenopus laevis)
|
|
chr6_+_56954801
|
3.808
|
NM_001031623
NM_015555
|
ZNF451
|
zinc finger protein 451
|
|
chr5_+_156820161
|
3.776
|
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr15_-_55562483
|
3.732
|
NM_183234
NM_004580
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr18_-_51690988
|
3.725
|
|
MBD2
|
methyl-CpG binding domain protein 2
|
|
chr11_-_35441501
|
3.710
|
NM_001195728
NM_001252652
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr22_+_40322609
|
3.671
|
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr4_+_175204827
|
3.661
|
NM_001040157
|
CEP44
|
centrosomal protein 44kDa
|
|
chr6_+_56954985
|
3.658
|
|
ZNF451
|
zinc finger protein 451
|
|
chr1_+_62902653
|
3.649
|
|
USP1
|
ubiquitin specific peptidase 1
|
|
chr4_-_83347778
|
3.629
|
|
HNRPDL
|
heterogeneous nuclear ribonucleoprotein D-like
|
|
chr12_-_102512277
|
3.611
|
NM_024057
|
NUP37
|
nucleoporin 37kDa
|
|
chr14_-_95599794
|
3.604
|
NM_001195573
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr3_-_148939784
|
3.589
|
NM_000096
|
CP
|
ceruloplasmin (ferroxidase)
|
|
chr21_-_40720938
|
3.572
|
|
HMGN1
|
high mobility group nucleosome binding domain 1
|
|
chr2_+_103035148
|
3.566
|
NM_003853
|
IL18RAP
|
interleukin 18 receptor accessory protein
|
|
chr13_-_48987247
|
3.560
|
|
LPAR6
|
lysophosphatidic acid receptor 6
|
|
chr4_-_83350829
|
3.556
|
|
HNRPDL
|
heterogeneous nuclear ribonucleoprotein D-like
|
|
chr4_-_103266577
|
3.542
|
NM_001135146
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8
|
|
chr14_-_35591151
|
3.540
|
|
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma
|
|
chr12_-_15114495
|
3.535
|
NM_001175
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta
|
|
chr5_+_102465256
|
3.517
|
NM_015216
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2
|
|
chr12_+_54674531
|
3.514
|
|
HNRNPA1L2
HNRNPA1
|
heterogeneous nuclear ribonucleoprotein A1-like 2
heterogeneous nuclear ribonucleoprotein A1
|
|
chr16_-_11375094
|
3.500
|
NM_002761
|
PRM1
|
protamine 1
|
|
chr12_-_24103842
|
3.498
|
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr1_+_84647342
|
3.488
|
NM_001242862
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr3_-_119582422
|
3.466
|
|
GSK3B
|
glycogen synthase kinase 3 beta
|
|
chr4_-_71532341
|
3.410
|
NM_144646
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides
|
|
chr8_+_26150731
|
3.401
|
NM_001177591
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chr2_+_86947387
|
3.366
|
NM_022780
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae)
|
|
chr6_+_15246299
|
3.349
|
NM_004973
|
JARID2
|
jumonji, AT rich interactive domain 2
|
|
chr2_-_70475563
|
3.322
|
NM_022037
NM_022173
|
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein
|
|
chr1_-_108231122
|
3.316
|
NM_001079874
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chrX_+_41192650
|
3.304
|
NM_001193416
NM_001193417
NM_001356
|
DDX3X
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked
|
|
chr6_-_79944397
|
3.300
|
NM_001201362
NM_001201363
NM_004242
NM_138730
|
HMGN3
|
high mobility group nucleosomal binding domain 3
|
|
chr2_-_88285308
|
3.287
|
NM_001024457
|
RGPD1
|
RANBP2-like and GRIP domain containing 1
|
|
chr2_+_10262694
|
3.282
|
NM_001165931
|
RRM2
|
ribonucleotide reductase M2
|
|
chr10_+_97803064
|
3.282
|
NM_001134375
NM_001134376
NM_019084
|
CCNJ
|
cyclin J
|
|
chr11_+_128563973
|
3.228
|
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr12_-_49319211
|
3.214
|
NM_001143782
NM_016594
|
FKBP11
|
FK506 binding protein 11, 19 kDa
|
|
chr11_-_16996559
|
3.211
|
|
RPL36A
|
ribosomal protein L36a
|
|
chr1_-_21503304
|
3.206
|
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3
|
|
chr13_-_48987017
|
3.197
|
|
LPAR6
|
lysophosphatidic acid receptor 6
|
|
chr1_-_53018721
|
3.191
|
NM_001009881
NM_015269
|
ZCCHC11
|
zinc finger, CCHC domain containing 11
|
|
chr12_+_14538232
|
3.175
|
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chr4_-_76598611
|
3.144
|
NM_203504
NM_203505
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2
|
|
chr8_+_103563847
|
3.143
|
NM_024410
|
ODF1
|
outer dense fiber of sperm tails 1
|
|
chr12_-_54653317
|
3.143
|
NM_001127322
|
CBX5
|
chromobox homolog 5
|
|
chrX_-_78623006
|
3.141
|
NM_001171581
NM_004867
|
ITM2A
|
integral membrane protein 2A
|
|
chr3_+_63884074
|
3.128
|
NM_001177387
|
ATXN7
|
ataxin 7
|
|
chr4_-_104020964
|
3.118
|
NM_020139
|
BDH2
|
3-hydroxybutyrate dehydrogenase, type 2
|
|
chr2_+_86947429
|
3.087
|
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae)
|
|
chr10_-_98290309
|
3.081
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr17_+_58677489
|
3.058
|
NM_003620
|
PPM1D
|
protein phosphatase, Mg2+/Mn2+ dependent, 1D
|
|
chr6_-_49604524
|
3.053
|
NM_000324
|
RHAG
|
Rh-associated glycoprotein
|
|
chr14_+_67656109
|
3.047
|
NM_173526
|
FAM71D
|
family with sequence similarity 71, member D
|
|
chr6_-_24719286
|
3.046
|
NM_030939
|
C6orf62
|
chromosome 6 open reading frame 62
|
|
chr6_+_100054649
|
3.043
|
NM_021620
|
PRDM13
|
PR domain containing 13
|
|
chr19_+_34710327
|
3.039
|
|
LSM14A
|
LSM14A, SCD6 homolog A (S. cerevisiae)
|
|
chr6_-_79944324
|
3.035
|
|
HMGN3
|
high mobility group nucleosomal binding domain 3
|
|
chr2_-_86422591
|
3.032
|
|
IMMT
|
inner membrane protein, mitochondrial
|
|
chr10_+_105727469
|
3.023
|
NM_014720
|
SLK
|
STE20-like kinase
|
|
chr2_+_232573249
|
3.014
|
|
PTMA
|
prothymosin, alpha
|
|
chr12_+_21525801
|
3.003
|
NM_000415
|
IAPP
|
islet amyloid polypeptide
|
|
chr3_-_57234279
|
3.002
|
NM_003865
|
HESX1
|
HESX homeobox 1
|
|
chr3_-_135913276
|
2.996
|
NM_001145417
|
MSL2
|
male-specific lethal 2 homolog (Drosophila)
|