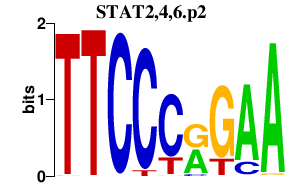
|
chr5_+_71502700
|
51.446
|
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr14_+_95078713
|
39.281
|
NM_001085
|
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3
|
|
chr12_-_9268522
|
32.247
|
NM_000014
|
A2M
|
alpha-2-macroglobulin
|
|
chr1_+_6845381
|
27.133
|
NM_001195563
NM_001242701
NM_015215
|
CAMTA1
|
calmodulin binding transcription activator 1
|
|
chr12_-_7245006
|
24.410
|
NM_001733
|
C1R
|
complement component 1, r subcomponent
|
|
chr12_-_7245048
|
24.165
|
|
C1R
|
complement component 1, r subcomponent
|
|
chr12_+_53443834
|
23.601
|
NM_170754
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr11_+_57365026
|
23.363
|
NM_000062
|
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1
|
|
chr11_+_57365577
|
22.334
|
NM_001032295
|
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1
|
|
chr12_+_58138663
|
19.063
|
NM_005981
|
TSPAN31
|
tetraspanin 31
|
|
chr8_-_120651019
|
18.991
|
NM_001040092
NM_001130863
NM_006209
|
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2
|
|
chr12_-_9268440
|
18.576
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr1_+_155853546
|
18.465
|
|
SYT11
|
synaptotagmin XI
|
|
chr11_+_61197574
|
17.939
|
NM_017841
|
SDHAF2
|
succinate dehydrogenase complex assembly factor 2
|
|
chr12_-_9268513
|
17.467
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr11_-_12030128
|
17.077
|
NM_001018057
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr3_+_159557745
|
16.909
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr1_-_230850335
|
16.583
|
NM_000029
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr12_+_133656984
|
15.707
|
NM_003440
|
ZNF140
|
zinc finger protein 140
|
|
chr1_-_230849863
|
15.647
|
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr15_+_71184781
|
15.582
|
NM_001199017
NM_017691
NM_001199018
|
LRRC49
|
leucine rich repeat containing 49
|
|
chr4_-_176734181
|
15.428
|
NM_201591
|
GPM6A
|
glycoprotein M6A
|
|
chr17_+_32582295
|
15.137
|
NM_002982
|
CCL2
|
chemokine (C-C motif) ligand 2
|
|
chr3_+_133495925
|
14.366
|
|
TF
|
transferrin
|
|
chr12_-_9268475
|
14.169
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr19_-_44123732
|
14.053
|
|
ZNF428
|
zinc finger protein 428
|
|
chr1_-_230849847
|
13.954
|
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr4_+_41258909
|
13.908
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr4_+_88896899
|
13.854
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr4_+_41258895
|
13.725
|
NM_004181
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr4_+_41258941
|
13.602
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr3_-_187463227
|
13.389
|
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr10_-_90750955
|
13.203
|
NM_001141945
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr11_-_12030622
|
13.179
|
NM_015881
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr4_+_88896868
|
13.120
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr20_+_44036940
|
13.011
|
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2
|
|
chr12_+_7167979
|
12.941
|
NM_001734
NM_201442
|
C1S
|
complement component 1, s subcomponent
|
|
chr19_-_37019169
|
12.906
|
NM_001012756
NM_001166036
NM_001166037
NM_001166038
|
ZNF260
|
zinc finger protein 260
|
|
chr11_-_12030539
|
12.655
|
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr4_+_88896801
|
12.605
|
NM_000582
NM_001040058
NM_001040060
NM_001251829
NM_001251830
|
SPP1
|
secreted phosphoprotein 1
|
|
chr1_+_169075869
|
12.528
|
NM_001677
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr4_+_88896838
|
12.353
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr5_-_138210992
|
12.127
|
|
CTNNA1
LRRTM2
|
catenin (cadherin-associated protein), alpha 1, 102kDa
leucine rich repeat transmembrane neuronal 2
|
|
chr5_+_172483285
|
12.062
|
NM_001168393
NM_001168394
NM_153607
|
C5orf41
|
chromosome 5 open reading frame 41
|
|
chr8_+_26371461
|
11.786
|
NM_001197293
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr19_+_10381516
|
11.737
|
NM_000201
|
ICAM1
|
intercellular adhesion molecule 1
|
|
chr5_-_151066445
|
11.719
|
|
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin)
|
|
chr5_-_151066492
|
11.685
|
NM_003118
|
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin)
|
|
chr19_-_37407079
|
11.650
|
NM_001037232
|
ZNF829
|
zinc finger protein 829
|
|
chr8_+_26435920
|
11.495
|
NM_001244604
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr2_+_233498206
|
11.411
|
NM_025202
|
EFHD1
|
EF-hand domain family, member D1
|
|
chr8_-_144241918
|
11.399
|
NM_001130478
|
LY6H
|
lymphocyte antigen 6 complex, locus H
|
|
chr5_+_140261792
|
11.350
|
NM_018904
NM_031865
|
PCDHA13
|
protocadherin alpha 13
|
|
chr19_+_10381763
|
11.180
|
|
ICAM1
|
intercellular adhesion molecule 1
|
|
chr11_+_73357222
|
11.072
|
NM_001130033
NM_021200
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr16_-_31085503
|
10.948
|
NM_024706
|
ZNF668
|
zinc finger protein 668
|
|
chr12_-_6798419
|
10.695
|
|
ZNF384
|
zinc finger protein 384
|
|
chr8_-_144241678
|
10.600
|
|
LY6H
|
lymphocyte antigen 6 complex, locus H
|
|
chr3_-_187463474
|
10.496
|
NM_001706
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr3_+_159557649
|
10.462
|
NM_001197109
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr6_-_111136616
|
10.460
|
|
CDK19
|
cyclin-dependent kinase 19
|
|
chr19_-_44860798
|
10.364
|
NM_001083335
NM_013380
|
ZFP112
|
zinc finger protein 112 homolog (mouse)
|
|
chr9_+_74526527
|
10.263
|
|
C9orf85
|
chromosome 9 open reading frame 85
|
|
chr8_-_48650703
|
10.193
|
NM_005195
|
CEBPD
|
CCAAT/enhancer binding protein (C/EBP), delta
|
|
chr19_+_44669214
|
10.078
|
NM_001146220
NM_001032373
NM_001032374
NM_015919
NM_001032372
|
ZNF226
|
zinc finger protein 226
|
|
chr10_+_17851342
|
9.897
|
NM_002438
|
MRC1
|
mannose receptor, C type 1
|
|
chr8_-_48650683
|
9.781
|
|
CEBPD
|
CCAAT/enhancer binding protein (C/EBP), delta
|
|
chr13_+_111134933
|
9.693
|
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr9_-_89562103
|
9.658
|
NM_002048
|
GAS1
|
growth arrest-specific 1
|
|
chr19_+_44556163
|
9.596
|
NM_013361
|
ZNF223
|
zinc finger protein 223
|
|
chr1_+_171810611
|
9.582
|
NM_001136127
NM_015569
|
DNM3
|
dynamin 3
|
|
chr7_-_150038689
|
9.566
|
NM_002889
|
RARRES2
|
retinoic acid receptor responder (tazarotene induced) 2
|
|
chr19_-_49339667
|
9.510
|
|
HSD17B14
|
hydroxysteroid (17-beta) dehydrogenase 14
|
|
chr22_-_30162990
|
9.491
|
|
|
|
|
chrX_-_13835313
|
9.490
|
NM_001001995
NM_001001996
NM_005278
|
GPM6B
|
glycoprotein M6B
|
|
chr5_+_40909558
|
9.429
|
NM_000587
|
C7
|
complement component 7
|
|
chr1_-_144994908
|
9.300
|
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr19_-_42746743
|
9.295
|
|
GSK3A
|
glycogen synthase kinase 3 alpha
|
|
chr5_+_40909617
|
9.289
|
|
C7
|
complement component 7
|
|
chr19_+_45418063
|
9.237
|
|
APOC1
|
apolipoprotein C-I
|
|
chr19_+_15218141
|
9.158
|
NM_033025
|
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans)
|
|
chr1_-_144994839
|
9.151
|
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr2_-_38303265
|
9.150
|
|
CYP1B1
|
cytochrome P450, family 1, subfamily B, polypeptide 1
|
|
chr3_+_14166543
|
9.126
|
|
TMEM43
|
transmembrane protein 43
|
|
chr14_-_21270537
|
9.083
|
NM_198232
|
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic)
|
|
chr15_+_90808833
|
9.021
|
NM_001033088
|
NGRN
|
neugrin, neurite outgrowth associated
|
|
chr3_+_115342150
|
8.957
|
NM_001130064
NM_002045
|
GAP43
|
growth associated protein 43
|
|
chr20_+_43991739
|
8.937
|
NM_001099791
NM_033542
|
SYS1-DBNDD2
SYS1
|
SYS1-DBNDD2 readthrough
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae)
|
|
chr2_+_17935379
|
8.876
|
NM_001130009
|
GEN1
|
Gen endonuclease homolog 1 (Drosophila)
|
|
chr3_+_14166582
|
8.822
|
|
TMEM43
|
transmembrane protein 43
|
|
chrX_-_13835012
|
8.812
|
|
GPM6B
|
glycoprotein M6B
|
|
chr8_-_26371387
|
8.806
|
NM_007257
|
PNMA2
|
paraneoplastic antigen MA2
|
|
chr19_+_54024176
|
8.744
|
NM_018555
|
ZNF331
|
zinc finger protein 331
|
|
chr14_-_60097223
|
8.742
|
|
RTN1
|
reticulon 1
|
|
chrX_+_102632108
|
8.669
|
NM_014380
|
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr20_+_44036628
|
8.631
|
NM_001048225
NM_001048226
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2
|
|
chr3_+_186330711
|
8.574
|
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr18_+_29171729
|
8.500
|
NM_000371
|
TTR
|
transthyretin
|
|
chr16_+_56659584
|
8.470
|
NM_175617
|
MT1E
|
metallothionein 1E
|
|
chr1_-_144994943
|
8.464
|
NM_001002812
NM_014644
NM_001002810
NM_001198834
NM_001195261
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr16_+_53738044
|
8.445
|
|
FTO
|
fat mass and obesity associated
|
|
chr3_+_186330844
|
8.438
|
NM_001622
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr8_-_144241725
|
8.391
|
NM_001135655
|
LY6H
|
lymphocyte antigen 6 complex, locus H
|
|
chr5_-_138211054
|
8.348
|
NM_015564
|
LRRTM2
|
leucine rich repeat transmembrane neuronal 2
|
|
chr17_-_1619473
|
8.313
|
|
MIR22HG
|
MIR22 host gene (non-protein coding)
|
|
chr3_+_115342302
|
8.295
|
|
GAP43
|
growth associated protein 43
|
|
chr22_+_29138018
|
8.280
|
NM_172002
|
HSCB
|
HscB iron-sulfur cluster co-chaperone homolog (E. coli)
|
|
chr19_-_44124013
|
8.195
|
NM_182498
|
ZNF428
|
zinc finger protein 428
|
|
chr12_+_13197284
|
8.194
|
NM_020853
|
KIAA1467
|
KIAA1467
|
|
chr7_+_86273951
|
8.192
|
|
GRM3
|
glutamate receptor, metabotropic 3
|
|
chr2_+_17935124
|
8.142
|
NM_182625
|
GEN1
|
Gen endonuclease homolog 1 (Drosophila)
|
|
chr3_+_186330859
|
8.129
|
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr17_-_29624249
|
8.093
|
NM_002544
|
OMG
|
oligodendrocyte myelin glycoprotein
|
|
chr19_+_44617510
|
8.089
|
NM_013362
|
ZNF225
|
zinc finger protein 225
|
|
chr7_-_37025696
|
8.020
|
|
ELMO1
|
engulfment and cell motility 1
|
|
chr11_+_114167168
|
7.869
|
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr3_+_14166439
|
7.864
|
NM_024334
|
TMEM43
|
transmembrane protein 43
|
|
chr15_-_71184611
|
7.813
|
NM_020147
|
THAP10
|
THAP domain containing 10
|
|
chr1_-_35497447
|
7.773
|
|
ZMYM6
|
zinc finger, MYM-type 6
|
|
chr10_+_88728187
|
7.726
|
NM_006829
|
C10orf116
|
chromosome 10 open reading frame 116
|
|
chr19_+_57791281
|
7.725
|
|
ZNF460
|
zinc finger protein 460
|
|
chr19_+_40502942
|
7.678
|
NM_178544
|
ZNF546
|
zinc finger protein 546
|
|
chr3_-_50649080
|
7.653
|
|
CISH
|
cytokine inducible SH2-containing protein
|
|
chr16_+_67204404
|
7.646
|
NM_001185058
|
NOL3
|
nucleolar protein 3 (apoptosis repressor with CARD domain)
|
|
chr1_-_183274008
|
7.601
|
NM_170706
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2
|
|
chr14_-_53417272
|
7.563
|
|
FERMT2
|
fermitin family member 2
|
|
chr19_-_42746678
|
7.535
|
|
GSK3A
|
glycogen synthase kinase 3 alpha
|
|
chr11_+_114167197
|
7.483
|
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr8_+_22462273
|
7.466
|
|
KIAA1967
|
KIAA1967
|
|
chr8_+_22462267
|
7.449
|
|
KIAA1967
|
KIAA1967
|
|
chr6_-_28304151
|
7.447
|
NM_001243241
NM_001243242
NM_001243244
NM_030899
|
ZNF323
|
zinc finger protein 323
|
|
chrX_-_48900932
|
7.420
|
NM_006521
|
TFE3
|
transcription factor binding to IGHM enhancer 3
|
|
chr1_+_36690038
|
7.395
|
|
THRAP3
|
thyroid hormone receptor associated protein 3
|
|
chr19_-_49339913
|
7.389
|
NM_016246
|
HSD17B14
|
hydroxysteroid (17-beta) dehydrogenase 14
|
|
chr12_+_12938540
|
7.387
|
NM_030817
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr7_+_86274076
|
7.381
|
|
GRM3
|
glutamate receptor, metabotropic 3
|
|
chr4_-_155511838
|
7.375
|
NM_000508
NM_021871
|
FGA
|
fibrinogen alpha chain
|
|
chr13_-_36705432
|
7.329
|
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr4_+_41362750
|
7.313
|
NM_001112717
NM_001112718
NM_014988
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr3_+_133464883
|
7.299
|
NM_001063
|
TF
|
transferrin
|
|
chr8_-_110661007
|
7.297
|
NM_001099747
NM_001099748
NM_001099749
NM_001099750
NM_017786
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr1_+_36689996
|
7.251
|
NM_005119
|
THRAP3
|
thyroid hormone receptor associated protein 3
|
|
chr11_-_117186918
|
7.240
|
NM_012104
NM_138971
NM_138972
NM_138973
|
BACE1
|
beta-site APP-cleaving enzyme 1
|
|
chr1_+_155099935
|
7.233
|
|
EFNA1
|
ephrin-A1
|
|
chr16_+_31085751
|
7.197
|
|
ZNF646
|
zinc finger protein 646
|
|
chr8_-_27472188
|
7.186
|
|
CLU
|
clusterin
|
|
chr16_-_28550227
|
7.185
|
|
NUPR1
|
nuclear protein, transcriptional regulator, 1
|
|
chr16_+_56659679
|
7.165
|
|
MT1E
|
metallothionein 1E
|
|
chrX_-_48931645
|
7.121
|
NM_007213
|
PRAF2
|
PRA1 domain family, member 2
|
|
chr7_+_86273219
|
7.094
|
NM_000840
|
GRM3
|
glutamate receptor, metabotropic 3
|
|
chr16_+_53737874
|
7.071
|
NM_001080432
|
FTO
|
fat mass and obesity associated
|
|
chr8_+_22462286
|
7.034
|
|
KIAA1967
|
KIAA1967
|
|
chr19_-_36980758
|
6.984
|
|
ZNF566
|
zinc finger protein 566
|
|
chr4_-_155533882
|
6.928
|
NM_000509
NM_021870
|
FGG
|
fibrinogen gamma chain
|
|
chr4_-_155533757
|
6.895
|
|
FGG
|
fibrinogen gamma chain
|
|
chr8_+_1952875
|
6.860
|
|
KBTBD11
|
kelch repeat and BTB (POZ) domain containing 11
|
|
chr1_-_35497544
|
6.852
|
|
ZMYM6
|
zinc finger, MYM-type 6
|
|
chr12_-_93964636
|
6.851
|
|
|
|
|
chr6_+_31913763
|
6.822
|
|
CFB
|
complement factor B
|
|
chr14_-_105715740
|
6.798
|
NM_001242789
|
BRF1
|
BRF1 homolog, subunit of RNA polymerase III transcription initiation factor IIIB (S. cerevisiae)
|
|
chr16_+_31085722
|
6.698
|
NM_014699
|
ZNF646
|
zinc finger protein 646
|
|
chr8_+_26435466
|
6.695
|
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr14_-_60097531
|
6.687
|
NM_206852
|
RTN1
|
reticulon 1
|
|
chr8_-_95274520
|
6.660
|
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr9_-_130186491
|
6.632
|
|
|
|
|
chr4_-_88450601
|
6.586
|
NM_001128310
NM_004684
|
SPARCL1
|
SPARC-like 1 (hevin)
|
|
chr19_-_42746723
|
6.526
|
NM_019884
|
GSK3A
|
glycogen synthase kinase 3 alpha
|
|
chr5_-_140013034
|
6.464
|
NM_000591
|
CD14
|
CD14 molecule
|
|
chr8_-_27472298
|
6.450
|
NM_001831
|
CLU
|
clusterin
|
|
chr16_-_28550324
|
6.449
|
NM_001042483
NM_012385
|
NUPR1
|
nuclear protein, transcriptional regulator, 1
|
|
chr3_-_139108486
|
6.449
|
|
|
|
|
chr17_-_42992852
|
6.441
|
NM_001131019
NM_001242376
NM_002055
|
GFAP
|
glial fibrillary acidic protein
|
|
chr4_-_155533850
|
6.440
|
|
FGG
|
fibrinogen gamma chain
|
|
chr19_+_48867650
|
6.402
|
NM_012451
|
SYNGR4
|
synaptogyrin 4
|
|
chr11_+_73358593
|
6.398
|
NM_001130034
NM_001130035
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr8_-_22089850
|
6.391
|
NM_001099335
NM_014759
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein
|
|
chr16_-_56701904
|
6.387
|
NM_005950
|
MT1G
|
metallothionein 1G
|
|
chr17_+_40834571
|
6.378
|
NM_003632
|
CNTNAP1
|
contactin associated protein 1
|
|
chr1_+_54519259
|
6.358
|
NM_153035
|
TCEANC2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2
|
|
chr12_-_45269340
|
6.352
|
NM_001145109
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr2_+_79347487
|
6.333
|
NM_002909
|
REG1A
|
regenerating islet-derived 1 alpha
|
|
chr14_+_94577078
|
6.333
|
NM_001130080
NM_005532
|
IFI27
|
interferon, alpha-inducible protein 27
|
|
chr17_+_76183397
|
6.329
|
NM_001010982
NM_001145526
|
AFMID
|
arylformamidase
|
|
chr5_-_140013285
|
6.297
|
NM_001040021
NM_001174104
NM_001174105
|
CD14
|
CD14 molecule
|
|
chr1_-_35497563
|
6.292
|
NM_007167
|
ZMYM6
|
zinc finger, MYM-type 6
|
|
chr13_-_36705513
|
6.288
|
NM_004734
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr4_-_155533800
|
6.285
|
|
FGG
|
fibrinogen gamma chain
|
|
chr2_+_17721806
|
6.279
|
NM_003385
|
VSNL1
|
visinin-like 1
|
|
chr8_+_22462532
|
6.274
|
NM_021174
|
KIAA1967
|
KIAA1967
|
|
chr2_+_31456879
|
6.272
|
NM_014600
|
EHD3
|
EH-domain containing 3
|
|
chr6_-_28303910
|
6.262
|
NM_001135216
NM_001243243
|
ZNF323
|
zinc finger protein 323
|
|
chr1_-_156217717
|
6.243
|
NM_198406
NM_024897
|
PAQR6
|
progestin and adipoQ receptor family member VI
|
|
chr17_+_74733468
|
6.242
|
NM_001242532
NM_001242533
NM_001242535
NM_001242536
NM_001242537
NM_024311
|
MFSD11
|
major facilitator superfamily domain containing 11
|
|
chr3_+_45067750
|
6.214
|
NM_003278
|
CLEC3B
|
C-type lectin domain family 3, member B
|
|
chr11_-_72380107
|
6.203
|
NM_001146209
|
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated
|
|
chr1_-_163113938
|
6.194
|
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr2_+_201450730
|
6.187
|
NM_001159
|
AOX1
|
aldehyde oxidase 1
|
|
chr12_-_6798528
|
6.186
|
NM_133476
|
ZNF384
|
zinc finger protein 384
|
|
chr12_-_6798619
|
6.170
|
NM_001039920
NM_001135734
|
ZNF384
|
zinc finger protein 384
|
|
chr19_+_56652689
|
6.163
|
|
ZNF444
|
zinc finger protein 444
|