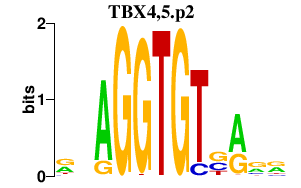
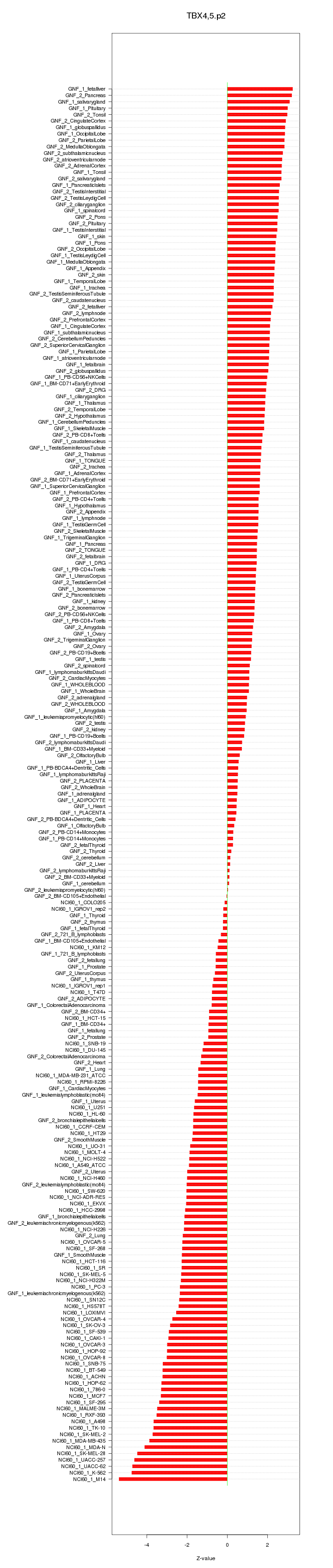
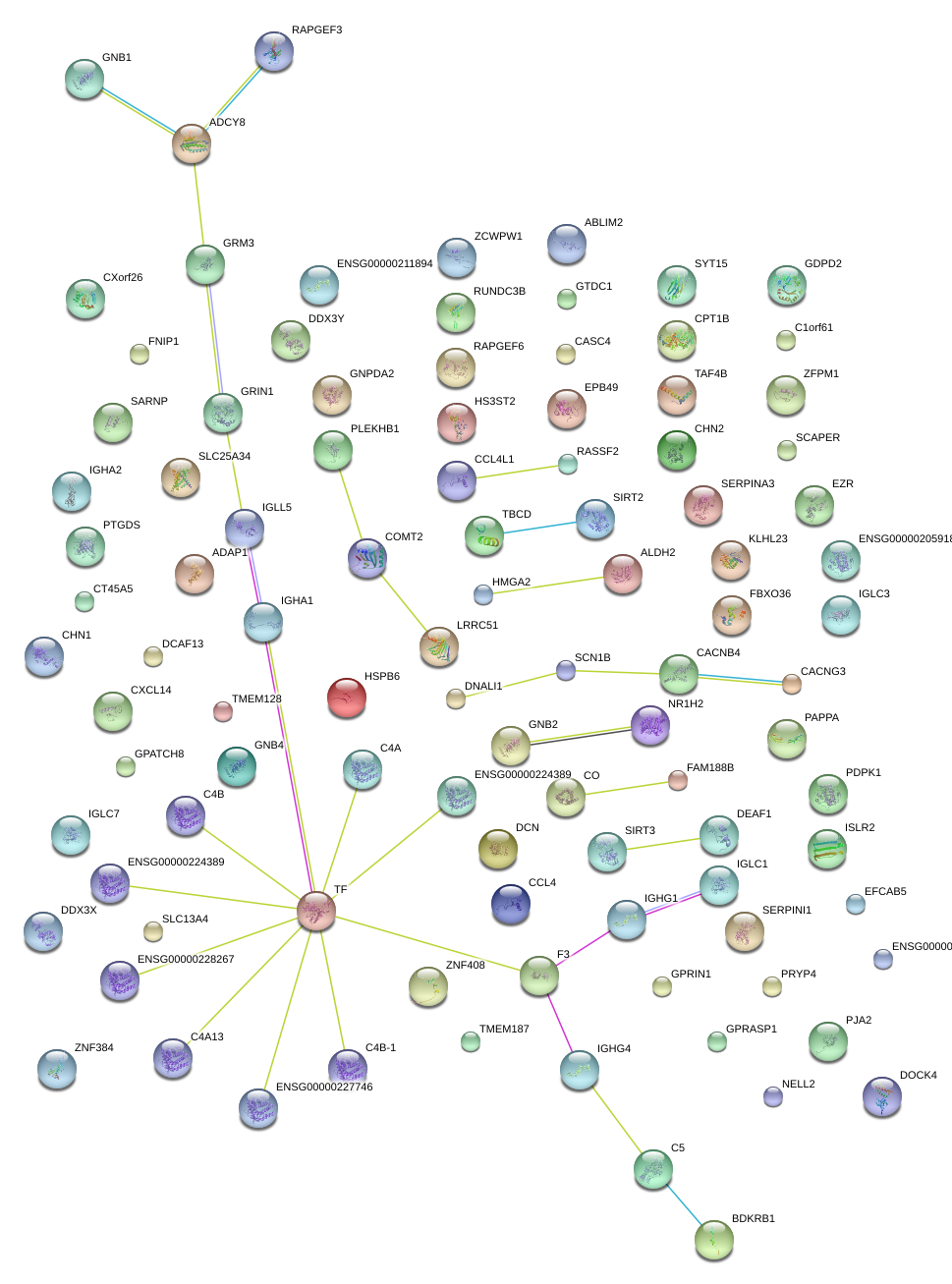
|
chr15_+_44581045
|
35.829
|
|
CASC4
|
cancer susceptibility candidate 4
|
|
chr15_+_74421986
|
32.746
|
NM_001130137
|
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2
|
|
chr3_+_167453431
|
29.133
|
NM_001122752
NM_005025
|
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1
|
|
chr9_+_139872014
|
28.805
|
|
PTGDS
|
prostaglandin D2 synthase 21kDa (brain)
|
|
chr15_+_74421698
|
28.790
|
NM_001130136
|
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2
|
|
chr12_-_45270071
|
27.480
|
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr1_+_156698270
|
25.018
|
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1
|
|
chr1_-_156399183
|
24.476
|
NM_006365
|
C1orf61
|
chromosome 1 open reading frame 61
|
|
chr15_+_44580954
|
24.113
|
|
CASC4
|
cancer susceptibility candidate 4
|
|
chr14_+_95078713
|
23.540
|
NM_001085
|
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3
|
|
chr12_+_66218879
|
23.422
|
|
HMGA2
|
high mobility group AT-hook 2
|
|
chr15_+_44580870
|
23.203
|
NM_138423
NM_177974
|
CASC4
|
cancer susceptibility candidate 4
|
|
chrX_+_75392763
|
23.112
|
NM_016500
|
CXorf26
|
chromosome X open reading frame 26
|
|
chr16_+_22825481
|
22.268
|
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2
|
|
chrX_+_101906293
|
21.641
|
NM_001099410
NM_001099411
NM_014710
NM_001184727
|
GPRASP1
|
G protein-coupled receptor associated sorting protein 1
|
|
chr11_+_71791376
|
21.375
|
NM_001145307
NM_001145308
NM_001205138
NM_145309
|
LRTOMT
|
leucine rich transmembrane and 0-methyltransferase domain containing
|
|
chrY_+_15016698
|
20.161
|
NM_004660
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked
|
|
chr1_+_156698250
|
20.005
|
NM_001142560
NM_015997
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1
|
|
chr5_-_131132642
|
19.992
|
|
FNIP1
|
folliculin interacting protein 1
|
|
chr12_-_45270157
|
19.500
|
NM_001145108
NM_006159
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr8_+_21912327
|
18.817
|
NM_001114136
NM_001114139
|
EPB49
|
erythrocyte membrane protein band 4.9 (dematin)
|
|
chr12_-_91576579
|
18.734
|
|
DCN
|
decorin
|
|
chr12_-_45269953
|
18.234
|
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr12_-_6798528
|
18.149
|
NM_133476
|
ZNF384
|
zinc finger protein 384
|
|
chr22_+_23229959
|
18.092
|
NM_001178126
|
IGLL5
IGLC1
|
immunoglobulin lambda-like polypeptide 5
immunoglobulin lambda constant 1 (Mcg marker)
|
|
chr1_+_16062808
|
18.051
|
NM_207348
|
SLC25A34
|
solute carrier family 25, member 34
|
|
chr15_-_77197743
|
17.838
|
|
SCAPER
|
S-phase cyclin A-associated protein in the ER
|
|
chr5_-_42811959
|
17.650
|
NM_001085486
NM_001093726
NM_005410
|
SEPP1
|
selenoprotein P, plasma, 1
|
|
chr4_-_8160435
|
17.013
|
NM_001130083
NM_001130084
NM_001130085
NM_001130086
NM_001130087
NM_001130088
NM_032432
|
ABLIM2
|
actin binding LIM protein family, member 2
|
|
chr2_-_175869910
|
16.874
|
NM_001822
NM_001025201
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr7_-_111846384
|
16.872
|
NM_014705
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr2_-_89165652
|
16.544
|
|
|
|
|
chr17_-_42580745
|
16.257
|
|
GPATCH8
|
G patch domain containing 8
|
|
chr17_+_28256873
|
16.228
|
NM_001145053
|
EFCAB5
|
EF-hand calcium binding domain 5
|
|
chr20_-_4804067
|
16.154
|
NM_014737
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr7_-_994008
|
16.072
|
|
ADAP1
|
ArfGAP with dual PH domains 1
|
|
chr10_-_46970595
|
15.873
|
NM_031912
NM_181519
|
SYT15
|
synaptotagmin XV
|
|
chr2_-_145090028
|
15.803
|
|
GTDC1
|
glycosyltransferase-like domain containing 1
|
|
chr19_-_36247917
|
15.801
|
NM_144617
|
HSPB6
|
heat shock protein, alpha-crystallin-related, B6
|
|
chr11_+_73358593
|
15.733
|
NM_001130034
NM_001130035
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr9_-_123837410
|
15.722
|
|
C5
|
complement component 5
|
|
chr5_-_130970851
|
15.694
|
|
RAPGEF6
|
Rap guanine nucleotide exchange factor (GEF) 6
|
|
chr7_-_994259
|
15.496
|
NM_006869
|
ADAP1
|
ArfGAP with dual PH domains 1
|
|
chr7_+_135347211
|
15.232
|
NM_001130929
|
SLC13A4
C7orf73
|
solute carrier family 13 (sodium/sulfate symporters), member 4
chromosome 7 open reading frame 73
|
|
chr4_-_44728518
|
14.761
|
NM_138335
|
GNPDA2
|
glucosamine-6-phosphate deaminase 2
|
|
chr5_-_130970907
|
14.629
|
NM_001164386
NM_001164387
NM_001164388
NM_001164389
NM_001164390
NM_016340
|
RAPGEF6
|
Rap guanine nucleotide exchange factor (GEF) 6
|
|
chr19_-_39390168
|
14.533
|
NM_012237
NM_001193286
NM_030593
|
SIRT2
|
sirtuin 2
|
|
chr5_-_108745569
|
14.410
|
NM_014819
|
PJA2
|
praja ring finger 2
|
|
chr12_-_91576805
|
14.145
|
NM_001920
|
DCN
|
decorin
|
|
chr3_+_133465098
|
14.118
|
|
TF
|
transferrin
|
|
chr1_+_38022514
|
14.040
|
NM_003462
|
DNALI1
|
dynein, axonemal, light intermediate chain 1
|
|
chr7_+_29234111
|
14.030
|
NM_004067
|
CHN2
|
chimerin (chimaerin) 2
|
|
chr7_-_100026284
|
13.867
|
NM_017984
|
ZCWPW1
|
zinc finger, CW type with PWWP domain 1
|
|
chr14_-_107113943
|
13.825
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr2_+_230787195
|
13.660
|
NM_174899
|
FBXO36
|
F-box protein 36
|
|
chr16_+_2587989
|
13.537
|
|
PDPK1
|
3-phosphoinositide dependent protein kinase-1
|
|
chr7_+_29234325
|
13.364
|
|
CHN2
|
chimerin (chimaerin) 2
|
|
chr19_+_507475
|
13.341
|
NM_033513
|
TPGS1
|
tubulin polyglutamylase complex subunit 1
|
|
chr16_+_24267474
|
13.313
|
|
CACNG3
|
calcium channel, voltage-dependent, gamma subunit 3
|
|
chr12_-_56211503
|
13.296
|
|
SARNP
|
SAP domain containing ribonucleoprotein
|
|
chr4_-_4249928
|
12.999
|
NM_032927
|
TMEM128
|
transmembrane protein 128
|
|
chr6_+_31982538
|
12.736
|
NM_001242823
NM_001252204
NM_007293
NM_001002029
|
LOC100293534
C4A
C4B
|
complement C4-B-like
complement component 4A (Rodgers blood group)
complement component 4B (Chido blood group)
|
|
chr22_-_51016343
|
12.556
|
NM_001145137
|
CPT1B
|
carnitine palmitoyltransferase 1B (muscle)
|
|
chr2_-_65454716
|
12.492
|
|
LOC729324
|
hCG1986447
|
|
chrX_+_153237990
|
12.489
|
NM_003492
|
TMEM187
|
transmembrane protein 187
|
|
chr7_+_87257704
|
12.456
|
NM_001134405
NM_001134406
NM_138290
|
RUNDC3B
|
RUN domain containing 3B
|
|
chr2_-_152955810
|
12.432
|
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chrY_+_15016018
|
12.338
|
NM_001122665
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked
|
|
chr17_+_34431219
|
12.307
|
NM_002984
|
CCL4
|
chemokine (C-C motif) ligand 4
|
|
chr16_+_24267672
|
12.268
|
|
CACNG3
|
calcium channel, voltage-dependent, gamma subunit 3
|
|
chr11_-_695010
|
12.197
|
|
DEAF1
|
deformed epidermal autoregulatory factor 1 (Drosophila)
|
|
chr12_-_48152180
|
12.141
|
NM_001098532
NM_006105
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3
|
|
chr19_+_50879681
|
12.132
|
NM_007121
|
NR1H2
|
nuclear receptor subfamily 1, group H, member 2
|
|
chr5_-_134914968
|
12.129
|
NM_004887
|
CXCL14
|
chemokine (C-X-C motif) ligand 14
|
|
chr7_+_86274076
|
12.014
|
|
GRM3
|
glutamate receptor, metabotropic 3
|
|
chr11_+_46722564
|
11.965
|
NM_001184751
|
ZNF408
|
zinc finger protein 408
|
|
chrX_+_69642880
|
11.896
|
NM_001171191
NM_001171192
NM_001171193
NM_017711
|
GDPD2
|
glycerophosphodiester phosphodiesterase domain containing 2
|
|
chr1_-_178062734
|
11.818
|
|
LOC100302401
|
uncharacterized LOC100302401
|
|
chr3_-_179169330
|
11.818
|
NM_021629
|
GNB4
|
guanine nucleotide binding protein (G protein), beta polypeptide 4
|
|
chr9_+_140032774
|
11.758
|
|
GRIN1
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1
|
|
chr18_+_23806385
|
11.726
|
NM_005640
|
TAF4B
|
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa
|
|
chr19_+_35521591
|
11.691
|
NM_001037
NM_199037
|
SCN1B
|
sodium channel, voltage-gated, type I, beta
|
|
chr2_+_170590454
|
11.683
|
|
KLHL23
|
kelch-like 23 (Drosophila)
|
|
chr12_-_45270632
|
11.627
|
NM_001145107
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr11_-_236833
|
11.548
|
|
SIRT3
|
sirtuin 3
|
|
chr2_-_145090038
|
11.545
|
NM_001164629
|
GTDC1
|
glycosyltransferase-like domain containing 1
|
|
chr7_+_30951414
|
11.444
|
NM_198098
|
AQP1
|
aquaporin 1 (Colton blood group)
|
|
chr17_-_7123368
|
11.437
|
NM_001365
|
DLG4
|
discs, large homolog 4 (Drosophila)
|
|
chr2_-_237172987
|
11.404
|
NM_212556
|
ASB18
|
ankyrin repeat and SOCS box containing 18
|
|
chr10_-_93392857
|
11.320
|
NM_005398
|
PPP1R3C
|
protein phosphatase 1, regulatory subunit 3C
|
|
chr18_+_11981457
|
11.318
|
|
IMPA2
|
inositol(myo)-1(or 4)-monophosphatase 2
|
|
chr16_+_2588000
|
11.280
|
|
PDPK1
|
3-phosphoinositide dependent protein kinase-1
|
|
chr10_-_93392744
|
11.264
|
|
PPP1R3C
|
protein phosphatase 1, regulatory subunit 3C
|
|
chr19_-_9731883
|
11.262
|
NM_152289
|
ZNF561
|
zinc finger protein 561
|
|
chr6_-_28303910
|
11.235
|
NM_001135216
NM_001243243
|
ZNF323
|
zinc finger protein 323
|
|
chrX_+_102470003
|
11.216
|
NM_001080425
NM_001127688
|
BEX4
|
brain expressed, X-linked 4
|
|
chr12_+_51318743
|
10.983
|
|
METTL7A
|
methyltransferase like 7A
|
|
chr22_-_51016845
|
10.974
|
NM_004377
NM_152246
NM_001145134
NM_001145135
NM_001145136
NM_152245
|
CPT1B
|
carnitine palmitoyltransferase 1B (muscle)
|
|
chr10_+_114135013
|
10.950
|
NM_203380
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr11_-_695069
|
10.708
|
|
DEAF1
|
deformed epidermal autoregulatory factor 1 (Drosophila)
|
|
chr18_+_55102777
|
10.669
|
NM_004852
|
ONECUT2
|
one cut homeobox 2
|
|
chr1_-_241520384
|
10.615
|
|
RGS7
|
regulator of G-protein signaling 7
|
|
chr6_+_31949800
|
10.609
|
NM_001242823
NM_001252204
NM_007293
NM_001002029
|
LOC100293534
C4A
C4B
|
complement C4-B-like
complement component 4A (Rodgers blood group)
complement component 4B (Chido blood group)
|
|
chr19_-_54327428
|
10.528
|
NM_144687
|
NLRP12
|
NLR family, pyrin domain containing 12
|
|
chr19_-_46295647
|
10.516
|
|
DMWD
|
dystrophia myotonica, WD repeat containing
|
|
chr1_-_11907741
|
10.510
|
NM_006172
|
NPPA
|
natriuretic peptide A
|
|
chr19_-_39108534
|
10.505
|
NM_001042600
NM_007181
|
MAP4K1
|
mitogen-activated protein kinase kinase kinase kinase 1
|
|
chr7_+_100612689
|
10.489
|
|
|
|
|
chr3_+_133465204
|
10.485
|
|
TF
|
transferrin
|
|
chr19_+_38755097
|
10.444
|
NM_001166103
NM_021102
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr19_-_36499583
|
10.441
|
NM_001039876
|
C19orf46
|
chromosome 19 open reading frame 46
|
|
chr5_+_54320083
|
10.384
|
NM_002104
|
GZMK
|
granzyme K (granzyme 3; tryptase II)
|
|
chr2_-_73053030
|
10.304
|
NM_015189
|
EXOC6B
|
exocyst complex component 6B
|
|
chr2_+_61108702
|
10.297
|
|
REL
|
v-rel reticuloendotheliosis viral oncogene homolog (avian)
|
|
chr1_+_151584514
|
10.288
|
NM_030918
|
SNX27
|
sorting nexin family member 27
|
|
chrX_+_8432870
|
10.218
|
NM_001001888
|
VCX3B
VCX3A
|
variable charge, X-linked 3B
variable charge, X-linked 3A
|
|
chr19_+_7828034
|
10.212
|
NM_001144904
NM_001144905
NM_001144906
NM_001144907
NM_001144908
NM_001144909
NM_001144910
NM_001144911
NM_014257
|
CLEC4M
|
C-type lectin domain family 4, member M
|
|
chr3_+_133464883
|
10.209
|
NM_001063
|
TF
|
transferrin
|
|
chr1_+_44412418
|
10.189
|
NM_014652
|
IPO13
|
importin 13
|
|
chr19_+_49838700
|
10.176
|
|
CD37
|
CD37 molecule
|
|
chr16_+_31271277
|
10.148
|
NM_000632
NM_001145808
|
ITGAM
|
integrin, alpha M (complement component 3 receptor 3 subunit)
|
|
chr1_-_241520467
|
10.029
|
NM_002924
|
RGS7
|
regulator of G-protein signaling 7
|
|
chr19_+_49838734
|
10.005
|
|
CD37
|
CD37 molecule
|
|
chr6_-_84937313
|
9.994
|
NM_014895
|
KIAA1009
|
KIAA1009
|
|
chr12_+_79258794
|
9.907
|
|
SYT1
|
synaptotagmin I
|
|
chr2_+_61108724
|
9.905
|
NM_002908
|
REL
|
v-rel reticuloendotheliosis viral oncogene homolog (avian)
|
|
chr17_+_30814708
|
9.901
|
|
|
|
|
chr3_-_133748828
|
9.813
|
NM_005630
|
SLCO2A1
|
solute carrier organic anion transporter family, member 2A1
|
|
chr8_-_27101240
|
9.806
|
|
STMN4
|
stathmin-like 4
|
|
chr1_+_118148506
|
9.781
|
NM_017709
|
FAM46C
|
family with sequence similarity 46, member C
|
|
chr16_+_31271311
|
9.775
|
|
ITGAM
|
integrin, alpha M (complement component 3 receptor 3 subunit)
|
|
chr11_-_75236418
|
9.767
|
NM_030792
|
GDPD5
|
glycerophosphodiester phosphodiesterase domain containing 5
|
|
chr11_-_64490654
|
9.753
|
NM_015080
NM_138732
|
NRXN2
|
neurexin 2
|
|
chr7_-_103629962
|
9.746
|
NM_005045
NM_173054
|
RELN
|
reelin
|
|
chr12_+_51818592
|
9.728
|
NM_001039960
NM_004858
|
SLC4A8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8
|
|
chrX_-_70331297
|
9.636
|
NM_000206
|
IL2RG
|
interleukin 2 receptor, gamma
|
|
chr19_+_38755238
|
9.632
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr16_+_50776012
|
9.609
|
NM_001042412
|
CYLD
|
cylindromatosis (turban tumor syndrome)
|
|
chr16_+_2588021
|
9.578
|
|
PDPK1
|
3-phosphoinositide dependent protein kinase-1
|
|
chr10_-_100206643
|
9.546
|
|
HPS1
|
Hermansky-Pudlak syndrome 1
|
|
chr11_-_59383616
|
9.529
|
NM_002556
|
OSBP
|
oxysterol binding protein
|
|
chr21_+_34398210
|
9.509
|
NM_005806
|
OLIG2
|
oligodendrocyte lineage transcription factor 2
|
|
chr11_-_46722139
|
9.509
|
|
ARHGAP1
|
Rho GTPase activating protein 1
|
|
chr1_+_17846818
|
9.501
|
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like
|
|
chr6_-_159420796
|
9.488
|
|
RSPH3
|
radial spoke 3 homolog (Chlamydomonas)
|
|
chr9_+_74526527
|
9.484
|
|
C9orf85
|
chromosome 9 open reading frame 85
|
|
chr4_-_99579540
|
9.483
|
NM_005723
|
TSPAN5
|
tetraspanin 5
|
|
chr2_+_176972005
|
9.433
|
NM_021192
|
HOXD11
|
homeobox D11
|
|
chr10_-_99393175
|
9.422
|
NM_178832
|
MORN4
|
MORN repeat containing 4
|
|
chr12_+_51818669
|
9.411
|
|
SLC4A8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8
|
|
chr11_-_129062092
|
9.385
|
NM_001142685
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chrX_-_48693934
|
9.367
|
NM_013271
|
PCSK1N
|
proprotein convertase subtilisin/kexin type 1 inhibitor
|
|
chr11_+_46958228
|
9.345
|
NM_001003676
NM_001003677
NM_001003678
NM_024113
|
C11orf49
|
chromosome 11 open reading frame 49
|
|
chr11_+_125774271
|
9.289
|
NM_013264
|
DDX25
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 25
|
|
chr1_+_35225341
|
9.273
|
NM_153212
|
GJB4
|
gap junction protein, beta 4, 30.3kDa
|
|
chr1_+_118148668
|
9.236
|
|
FAM46C
|
family with sequence similarity 46, member C
|
|
chr19_+_50706868
|
9.221
|
NM_001077186
NM_001145809
NM_024729
|
MYH14
|
myosin, heavy chain 14, non-muscle
|
|
chr11_+_1889902
|
9.181
|
NM_001013253
|
LSP1
|
lymphocyte-specific protein 1
|
|
chr10_+_135192694
|
9.159
|
NM_152911
NM_207127
NM_207128
|
PAOX
|
polyamine oxidase (exo-N4-amino)
|
|
chr13_-_45150391
|
9.105
|
|
TSC22D1
|
TSC22 domain family, member 1
|
|
chr10_-_99531712
|
9.075
|
NM_003015
|
SFRP5
|
secreted frizzled-related protein 5
|
|
chr11_+_46722316
|
9.048
|
NM_024741
|
ZNF408
|
zinc finger protein 408
|
|
chr5_-_39203061
|
9.030
|
|
FYB
|
FYN binding protein
|
|
chr14_-_107013474
|
9.027
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr12_-_6579754
|
9.010
|
NM_014231
NM_016830
NM_199245
|
VAMP1
|
vesicle-associated membrane protein 1 (synaptobrevin 1)
|
|
chr19_-_5622794
|
8.919
|
NM_014649
|
SAFB2
|
scaffold attachment factor B2
|
|
chr3_-_133748727
|
8.900
|
|
SLCO2A1
|
solute carrier organic anion transporter family, member 2A1
|
|
chr17_+_14204326
|
8.900
|
NM_006041
|
HS3ST3B1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1
|
|
chr1_+_156589085
|
8.815
|
NM_021817
|
HAPLN2
|
hyaluronan and proteoglycan link protein 2
|
|
chr2_+_242127923
|
8.800
|
NM_001001666
NM_001001891
|
ANO7
|
anoctamin 7
|
|
chr19_+_49838673
|
8.792
|
NM_001040031
NM_001774
|
CD37
|
CD37 molecule
|
|
chr9_+_124048869
|
8.760
|
|
GSN
|
gelsolin
|
|
chr14_-_105886075
|
8.753
|
|
LOC100507437
|
uncharacterized LOC100507437
|
|
chr1_-_156786500
|
8.723
|
NM_001161441
NM_001161442
NM_001161444
NM_003975
|
SH2D2A
|
SH2 domain containing 2A
|
|
chr15_+_68871285
|
8.714
|
NM_006091
|
CORO2B
|
coronin, actin binding protein, 2B
|
|
chr19_+_2096930
|
8.664
|
|
IZUMO4
|
IZUMO family member 4
|
|
chr6_-_6320875
|
8.656
|
|
F13A1
|
coagulation factor XIII, A1 polypeptide
|
|
chr1_-_202129118
|
8.634
|
NM_080588
|
PTPN7
|
protein tyrosine phosphatase, non-receptor type 7
|
|
chr10_-_100206698
|
8.605
|
NM_000195
NM_182639
|
HPS1
|
Hermansky-Pudlak syndrome 1
|
|
chr1_-_169862982
|
8.580
|
|
SCYL3
|
SCY1-like 3 (S. cerevisiae)
|
|
chr5_-_134914447
|
8.566
|
|
CXCL14
|
chemokine (C-X-C motif) ligand 14
|
|
chr7_-_6312137
|
8.562
|
NM_004227
|
CYTH3
|
cytohesin 3
|
|
chr12_+_69742130
|
8.517
|
NM_000239
|
LYZ
|
lysozyme
|
|
chr11_+_61197574
|
8.489
|
NM_017841
|
SDHAF2
|
succinate dehydrogenase complex assembly factor 2
|
|
chr19_+_39833132
|
8.485
|
|
SAMD4B
|
sterile alpha motif domain containing 4B
|
|
chr1_-_85725315
|
8.436
|
NM_198077
|
C1orf52
|
chromosome 1 open reading frame 52
|
|
chr19_-_57183122
|
8.434
|
NM_001005850
|
ZNF835
|
zinc finger protein 835
|
|
chr8_+_21915371
|
8.401
|
NM_001114135
|
EPB49
|
erythrocyte membrane protein band 4.9 (dematin)
|
|
chr17_-_39968823
|
8.399
|
|
LEPREL4
|
leprecan-like 4
|
|
chr7_-_37025696
|
8.353
|
|
ELMO1
|
engulfment and cell motility 1
|
|
chr9_+_72873985
|
8.342
|
|
SMC5
|
structural maintenance of chromosomes 5
|
|
chr9_-_136024522
|
8.336
|
NM_001042368
|
RALGDS
|
ral guanine nucleotide dissociation stimulator
|
|
chr5_-_149669400
|
8.294
|
NM_015981
NM_171825
|
CAMK2A
|
calcium/calmodulin-dependent protein kinase II alpha
|
|
chr16_+_50775960
|
8.292
|
NM_001042355
NM_015247
|
CYLD
|
cylindromatosis (turban tumor syndrome)
|
|
chr9_-_35115844
|
8.286
|
NM_025182
|
FAM214B
|
family with sequence similarity 214, member B
|
|
chr11_-_71791734
|
8.279
|
|
NUMA1
|
nuclear mitotic apparatus protein 1
|
|
chr11_-_33891258
|
8.278
|
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr4_-_57547845
|
8.244
|
NM_001145460
NM_032495
NM_139212
|
HOPX
|
HOP homeobox
|
|
chr12_+_69742140
|
8.226
|
|
LYZ
|
lysozyme
|
|
chr15_-_65412175
|
8.222
|
|
PDCD7
|
programmed cell death 7
|