|
chr10_-_90712510
|
114.932
|
NM_001613
|
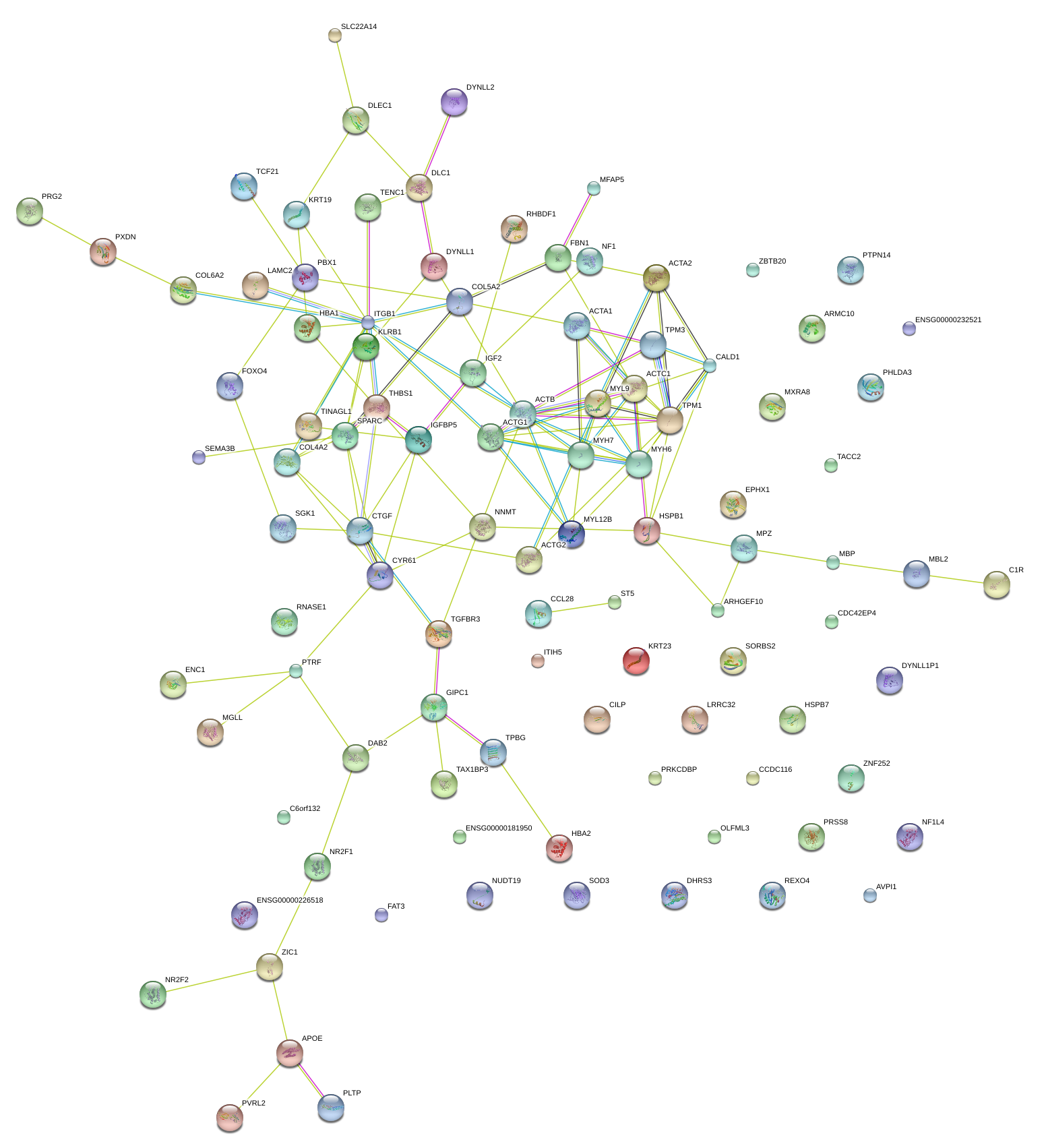
ACTA2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr6_-_132272311
|
108.515
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chr5_-_39424930
|
61.068
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr20_+_35169876
|
60.486
|
NM_006097
NM_181526
|
MYL9
|
myosin, light chain 9, regulatory
|
|
chr20_+_35169897
|
57.985
|
|
MYL9
|
myosin, light chain 9, regulatory
|
|
chr5_-_151066445
|
55.770
|
|
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin)
|
|
chr5_-_151066492
|
49.314
|
NM_003118
|
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin)
|
|
chr16_+_226678
|
46.493
|
NM_000558
|
HBA1
HBA2
|
hemoglobin, alpha 1
hemoglobin, alpha 2
|
|
chr2_-_217540722
|
45.776
|
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr16_+_222845
|
45.303
|
NM_000517
|
HBA1
HBA2
|
hemoglobin, alpha 1
hemoglobin, alpha 2
|
|
chr5_-_39425030
|
44.064
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr12_-_7245006
|
42.268
|
NM_001733
|
C1R
|
complement component 1, r subcomponent
|
|
chr12_-_7245048
|
41.904
|
|
C1R
|
complement component 1, r subcomponent
|
|
chr1_-_1293907
|
40.646
|
NM_032348
|
MXRA8
|
matrix-remodelling associated 8
|
|
chr15_+_63334753
|
39.510
|
NM_000366
NM_001018004
NM_001018005
NM_001018006
NM_001018007
NM_001018020
|
TPM1
|
tropomyosin 1 (alpha)
|
|
chr19_+_45409003
|
38.095
|
NM_000041
|
APOE
|
apolipoprotein E
|
|
chr1_-_12677347
|
37.446
|
NM_004753
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3
|
|
chr22_+_21986968
|
37.079
|
NM_152612
|
CCDC116
|
coiled-coil domain containing 116
|
|
chr15_+_63335086
|
35.987
|
|
TPM1
|
tropomyosin 1 (alpha)
|
|
chr11_+_114166534
|
34.727
|
NM_006169
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr11_-_6341708
|
33.695
|
NM_145040
|
PRKCDBP
|
protein kinase C, delta binding protein
|
|
chr3_-_114343052
|
32.923
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr14_-_21270537
|
31.615
|
NM_198232
|
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic)
|
|
chr4_-_186733372
|
30.742
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr20_-_44540685
|
30.695
|
|
PLTP
|
phospholipid transfer protein
|
|
chr15_+_39873276
|
30.315
|
NM_003246
|
THBS1
|
thrombospondin 1
|
|
chr17_-_40575117
|
30.106
|
NM_012232
|
PTRF
|
polymerase I and transcript release factor
|
|
chr5_-_39425297
|
28.839
|
NM_001244871
NM_001343
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr19_+_37288408
|
27.220
|
|
LOC284408
|
uncharacterized LOC284408
|
|
chr14_-_21271002
|
27.182
|
NM_002933
NM_198234
NM_198235
|
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic)
|
|
chr3_+_50306305
|
26.518
|
|
SEMA3B
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B
|
|
chr11_-_8832881
|
26.403
|
NM_139157
|
ST5
|
suppression of tumorigenicity 5
|
|
chr7_+_139877154
|
24.837
|
|
LOC100134229
|
uncharacterized LOC100134229
|
|
chr7_+_134464161
|
24.501
|
NM_004342
NM_033138
NM_033157
|
CALD1
|
caldesmon 1
|
|
chr7_+_134464417
|
22.559
|
|
CALD1
|
caldesmon 1
|
|
chr3_+_50306578
|
22.197
|
|
SEMA3B
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B
|
|
chr4_-_186578122
|
22.115
|
NM_001145674
NM_001145675
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr12_+_53442732
|
22.025
|
NM_015319
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr1_+_164528586
|
22.011
|
NM_001204961
NM_001204963
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr21_+_47545189
|
21.783
|
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr11_-_8832189
|
21.638
|
NM_213618
|
ST5
|
suppression of tumorigenicity 5
|
|
chr2_-_190044330
|
21.014
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr18_-_74728963
|
20.859
|
|
MBP
|
myelin basic protein
|
|
chr6_+_83073501
|
20.835
|
|
TPBG
|
trophoblast glycoprotein
|
|
chr11_-_2170792
|
20.535
|
NM_001007139
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr1_+_114522029
|
20.509
|
NM_020190
|
OLFML3
|
olfactomedin-like 3
|
|
chr6_+_134210247
|
20.426
|
NM_198392
NM_003206
|
TCF21
|
transcription factor 21
|
|
chr6_+_134210264
|
20.288
|
|
TCF21
|
transcription factor 21
|
|
chr10_-_99446899
|
20.145
|
NM_021732
|
AVPI1
|
arginine vasopressin-induced 1
|
|
chr13_+_111138079
|
20.144
|
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr1_-_214724844
|
20.026
|
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr10_+_123872440
|
19.919
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr17_-_39093671
|
19.852
|
NM_015515
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr7_+_102715580
|
19.779
|
|
ARMC10
|
armadillo repeat containing 10
|
|
chr1_+_32043745
|
19.558
|
NM_001204415
|
TINAGL1
|
tubulointerstitial nephritis antigen-like 1
|
|
chr4_+_24797203
|
19.489
|
|
SOD3
|
superoxide dismutase 3, extracellular
|
|
chr16_-_122573
|
19.361
|
NM_022450
|
RHBDF1
|
rhomboid 5 homolog 1 (Drosophila)
|
|
chr2_-_65454716
|
19.306
|
|
LOC729324
|
hCG1986447
|
|
chr1_+_226016428
|
18.836
|
|
EPHX1
|
epoxide hydrolase 1, microsomal (xenobiotic)
|
|
chr16_-_31146992
|
18.775
|
|
PRSS8
|
protease, serine, 8
|
|
chr6_+_83072922
|
18.520
|
NM_006670
|
TPBG
|
trophoblast glycoprotein
|
|
chr1_-_161279724
|
18.498
|
NM_000530
|
MPZ
|
myelin protein zero
|
|
chr1_-_16344438
|
18.101
|
|
HSPB7
|
heat shock 27kDa protein family, member 7 (cardiovascular)
|
|
chr15_-_48937795
|
17.901
|
NM_000138
|
FBN1
|
fibrillin 1
|
|
chr11_+_92085261
|
17.797
|
NM_001008781
|
FAT3
|
FAT tumor suppressor homolog 3 (Drosophila)
|
|
chr1_+_86046434
|
17.634
|
NM_001554
|
CYR61
|
cysteine-rich, angiogenic inducer, 61
|
|
chr10_-_33247178
|
17.553
|
NM_002211
|
ITGB1
|
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12)
|
|
chr6_-_134496833
|
16.925
|
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr3_+_147127151
|
16.510
|
NM_003412
|
ZIC1
|
Zic family member 1
|
|
chr1_-_92351613
|
16.439
|
NM_001195683
NM_003243
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr1_+_183155393
|
16.395
|
|
LAMC2
|
laminin, gamma 2
|
|
chr14_-_23904849
|
16.389
|
NM_000257
|
MYH7
|
myosin, heavy chain 7, cardiac muscle, beta
|
|
chr19_-_14606893
|
16.296
|
|
GIPC1
|
GIPC PDZ domain containing family, member 1
|
|
chr19_-_14606939
|
16.201
|
NM_005716
NM_202467
NM_202468
NM_202469
NM_202470
NM_202494
|
GIPC1
|
GIPC PDZ domain containing family, member 1
|
|
chr6_-_42110714
|
16.159
|
NM_001164446
|
C6orf132
|
chromosome 6 open reading frame 132
|
|
chr3_-_127541160
|
16.089
|
NM_007283
|
MGLL
|
monoglyceride lipase
|
|
chr7_+_102715293
|
15.813
|
NM_001161009
NM_001161010
NM_001161011
NM_001161012
NM_001161013
NM_031905
|
ARMC10
|
armadillo repeat containing 10
|
|
chr5_-_43412417
|
15.727
|
NM_148672
|
CCL28
|
chemokine (C-C motif) ligand 28
|
|
chr17_-_3571819
|
15.535
|
|
TAX1BP3
|
Tax1 (human T-cell leukemia virus type I) binding protein 3
|
|
chr10_-_7682741
|
15.350
|
|
ITIH5
|
inter-alpha-trypsin inhibitor heavy chain family, member 5
|
|
chr1_-_92351476
|
15.120
|
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr1_-_201438150
|
15.062
|
NM_012396
|
PHLDA3
|
pleckstrin homology-like domain, family A, member 3
|
|
chr19_+_45349584
|
14.758
|
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B)
|
|
chr6_-_134497069
|
14.357
|
NM_001143678
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr3_+_38347426
|
14.194
|
NM_004803
|
SLC22A14
|
solute carrier family 22, member 14
|
|
chr15_+_96869156
|
14.057
|
NM_001145155
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr8_-_12973752
|
13.855
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr12_-_8815370
|
13.774
|
NM_003480
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr19_+_33182689
|
13.761
|
NM_001105570
|
NUDT19
|
nudix (nucleoside diphosphate linked moiety X)-type motif 19
|
|
chr11_-_76380894
|
13.738
|
NM_005512
|
LRRC32
|
leucine rich repeat containing 32
|
|
chr16_-_31147062
|
13.625
|
NM_002773
|
PRSS8
|
protease, serine, 8
|
|
chr17_-_39684499
|
13.540
|
|
KRT19
|
keratin 19
|
|
chr17_+_29421956
|
13.309
|
|
NF1
|
neurofibromin 1
|
|
chr15_-_65503786
|
13.157
|
NM_003613
|
CILP
|
cartilage intermediate layer protein, nucleotide pyrophosphohydrolase
|
|
chrX_+_70315637
|
13.080
|
NM_001170931
NM_005938
|
FOXO4
|
forkhead box O4
|
|
chr8_+_1772095
|
12.882
|
NM_014629
|
ARHGEF10
|
Rho guanine nucleotide exchange factor (GEF) 10
|
|
chr7_+_75931829
|
12.810
|
NM_001540
|
HSPB1
|
heat shock 27kDa protein 1
|
|
chr8_-_146202727
|
12.799
|
|
ZNF252
|
zinc finger protein 252
|
|
chr10_-_33247131
|
12.797
|
|
ITGB1
|
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12)
|
|
chr17_-_71308118
|
12.764
|
NM_012121
|
CDC42EP4
|
CDC42 effector protein (Rho GTPase binding) 4
|
|
chr1_-_201346804
|
12.589
|
NM_000364
NM_001001430
NM_001001431
NM_001001432
|
TNNT2
|
troponin T type 2 (cardiac)
|
|
chr11_+_66824285
|
12.534
|
NM_014578
|
RHOD
|
ras homolog gene family, member D
|
|
chr7_+_134464372
|
12.448
|
|
CALD1
|
caldesmon 1
|
|
chrX_+_48620153
|
12.363
|
NM_001080489
|
GLOD5
|
glyoxalase domain containing 5
|
|
chr1_+_54519259
|
12.217
|
NM_153035
|
TCEANC2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2
|
|
chr12_-_8815479
|
11.988
|
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr12_-_8815266
|
11.895
|
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr7_+_116164838
|
11.795
|
NM_001753
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr7_-_5553401
|
11.571
|
|
FBXL18
|
F-box and leucine-rich repeat protein 18
|
|
chr8_-_27630096
|
11.459
|
NM_018246
|
CCDC25
|
coiled-coil domain containing 25
|
|
chr17_-_39684585
|
11.447
|
NM_002276
|
KRT19
|
keratin 19
|
|
chr15_-_60690115
|
11.329
|
NM_001002857
NM_001002858
NM_001136015
NM_004039
|
ANXA2
|
annexin A2
|
|
chr8_-_42065037
|
11.317
|
|
PLAT
|
plasminogen activator, tissue
|
|
chr19_+_36359346
|
11.273
|
NM_001024807
NM_005166
|
APLP1
|
amyloid beta (A4) precursor-like protein 1
|
|
chr11_-_5255712
|
11.214
|
NM_000519
|
HBD
|
hemoglobin, delta
|
|
chr8_-_42065193
|
11.168
|
NM_000930
NM_033011
|
PLAT
|
plasminogen activator, tissue
|
|
chr19_-_13617037
|
11.163
|
NM_000068
NM_001127221
NM_001127222
NM_001174080
NM_023035
|
CACNA1A
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit
|
|
chr17_+_48049678
|
10.968
|
|
DLX4
|
distal-less homeobox 4
|
|
chr19_-_49371710
|
10.957
|
NM_001161354
NM_020904
|
PLEKHA4
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 4
|
|
chr11_+_308106
|
10.864
|
NM_006435
|
IFITM2
|
interferon induced transmembrane protein 2 (1-8D)
|
|
chr7_-_102715171
|
10.708
|
NM_001111038
|
FBXL13
|
F-box and leucine-rich repeat protein 13
|
|
chr5_+_140868721
|
10.638
|
NM_018929
NM_032407
|
PCDHGC5
|
protocadherin gamma subfamily C, 5
|
|
chr1_-_214724974
|
10.610
|
NM_005401
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr1_-_115238091
|
10.541
|
NM_000036
NM_001172626
|
AMPD1
|
adenosine monophosphate deaminase 1
|
|
chr3_-_9994057
|
10.483
|
NM_207351
|
PRRT3
|
proline-rich transmembrane protein 3
|
|
chr17_-_73663221
|
10.473
|
NM_001003715
NM_001003716
NM_004259
|
RECQL5
|
RecQ protein-like 5
|
|
chr11_-_62521533
|
10.265
|
NM_024784
|
ZBTB3
|
zinc finger and BTB domain containing 3
|
|
chr4_-_57547845
|
10.249
|
NM_001145460
NM_032495
NM_139212
|
HOPX
|
HOP homeobox
|
|
chr11_-_6643809
|
10.213
|
|
DCHS1
|
dachsous 1 (Drosophila)
|
|
chr12_+_112856756
|
10.141
|
|
PTPN11
|
protein tyrosine phosphatase, non-receptor type 11
|
|
chr14_+_95047730
|
10.141
|
NM_000624
|
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5
|
|
chr17_+_37856253
|
10.114
|
NM_004448
|
ERBB2
|
v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian)
|
|
chr3_-_149086884
|
10.097
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr3_-_134093235
|
10.085
|
NM_016201
|
AMOTL2
|
angiomotin like 2
|
|
chr1_-_201346783
|
10.082
|
|
TNNT2
|
troponin T type 2 (cardiac)
|
|
chr4_-_186577858
|
9.989
|
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr11_+_66824308
|
9.974
|
|
RHOD
|
ras homolog gene family, member D
|
|
chr19_+_50936159
|
9.764
|
NM_004533
|
MYBPC2
|
myosin binding protein C, fast type
|
|
chr10_-_123353771
|
9.754
|
NM_001144916
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr19_+_44645708
|
9.723
|
NM_001144824
NM_006630
|
ZNF234
|
zinc finger protein 234
|
|
chr17_-_79105747
|
9.690
|
NM_004920
|
AATK
|
apoptosis-associated tyrosine kinase
|
|
chr7_-_102715014
|
9.541
|
NM_145032
|
FBXL13
|
F-box and leucine-rich repeat protein 13
|
|
chr11_+_71639767
|
9.520
|
NM_018320
|
RNF121
|
ring finger protein 121
|
|
chr5_+_34687663
|
9.339
|
NM_001145523
|
RAI14
|
retinoic acid induced 14
|
|
chr8_-_82395401
|
9.285
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr17_-_8079607
|
9.241
|
NM_032354
NM_183065
|
TMEM107
|
transmembrane protein 107
|
|
chr17_+_28268622
|
9.208
|
NM_198529
|
EFCAB5
|
EF-hand calcium binding domain 5
|
|
chr1_+_183155361
|
9.167
|
|
LAMC2
|
laminin, gamma 2
|
|
chr12_+_27677044
|
9.153
|
NM_001198915
NM_001198916
NM_003622
NM_177444
|
PPFIBP1
|
PTPRF interacting protein, binding protein 1 (liprin beta 1)
|
|
chr12_-_54813010
|
9.138
|
NM_002205
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr17_-_4693595
|
9.049
|
|
LOC284014
|
uncharacterized LOC284014
|
|
chr15_+_41136622
|
8.929
|
NM_001032367
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1
|
|
chr15_+_41136178
|
8.741
|
NM_003710
NM_181642
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1
|
|
chr19_+_11998609
|
8.719
|
NM_021915
|
ZNF69
|
zinc finger protein 69
|
|
chr3_-_52488030
|
8.678
|
NM_003280
|
TNNC1
|
troponin C type 1 (slow)
|
|
chr6_+_30850693
|
8.661
|
|
DDR1
|
discoidin domain receptor tyrosine kinase 1
|
|
chr19_+_36249043
|
8.599
|
NM_001039887
|
C19orf55
|
chromosome 19 open reading frame 55
|
|
chr10_-_44063906
|
8.510
|
NM_005674
|
ZNF239
|
zinc finger protein 239
|
|
chr4_+_184020343
|
8.468
|
NM_024949
|
WWC2
|
WW and C2 domain containing 2
|
|
chr10_-_69835012
|
8.457
|
|
HERC4
|
hect domain and RLD 4
|
|
chr16_-_1280162
|
8.456
|
NM_024164
|
TPSB2
TPSAB1
|
tryptase beta 2 (gene/pseudogene)
tryptase alpha/beta 1
|
|
chr3_-_49314281
|
8.437
|
NM_198562
|
C3orf62
|
chromosome 3 open reading frame 62
|
|
chr2_-_220407818
|
8.390
|
NM_001195731
|
CHPF
|
chondroitin polymerizing factor
|
|
chr4_+_176986984
|
8.373
|
NM_170710
NM_181265
|
WDR17
|
WD repeat domain 17
|
|
chr3_-_14989399
|
8.291
|
|
LOC100505641
|
uncharacterized LOC100505641
|
|
chr16_+_69984809
|
8.153
|
NM_182619
|
CLEC18C
CLEC18A
|
C-type lectin domain family 18, member C
C-type lectin domain family 18, member A
|
|
chr10_+_123748688
|
8.056
|
NM_206861
NM_206862
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr17_-_48278874
|
8.042
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr16_-_62070738
|
8.007
|
NM_001796
|
CDH8
|
cadherin 8, type 2
|
|
chr12_-_56101465
|
7.988
|
|
ITGA7
|
integrin, alpha 7
|
|
chr2_-_68384575
|
7.959
|
NM_138458
|
WDR92
|
WD repeat domain 92
|
|
chr1_+_19923457
|
7.929
|
NM_001204088
NM_001204089
NM_001032363
NM_001204082
NM_001204083
|
C1orf151-NBL1
MINOS1
|
C1orf151-NBL1 readthrough
mitochondrial inner membrane organizing system 1
|
|
chr17_+_29421941
|
7.913
|
NM_000267
NM_001042492
NM_001128147
|
NF1
|
neurofibromin 1
|
|
chr8_-_37727937
|
7.881
|
|
RAB11FIP1
|
RAB11 family interacting protein 1 (class I)
|
|
chr19_-_51504799
|
7.852
|
NM_007196
NM_144505
NM_144506
NM_144507
|
KLK8
|
kallikrein-related peptidase 8
|
|
chr20_+_47662873
|
7.753
|
|
CSE1L
|
CSE1 chromosome segregation 1-like (yeast)
|
|
chr12_-_56101667
|
7.634
|
NM_001144996
NM_002206
|
ITGA7
|
integrin, alpha 7
|
|
chr18_+_32173281
|
7.601
|
NM_001128175
NM_001198938
NM_001198939
NM_001198940
|
DTNA
|
dystrobrevin, alpha
|
|
chr1_+_20396652
|
7.600
|
NM_000929
|
PLA2G5
|
phospholipase A2, group V
|
|
chr2_-_238499317
|
7.565
|
|
RAB17
|
RAB17, member RAS oncogene family
|
|
chr21_-_27423423
|
7.528
|
|
|
|
|
chr6_+_123109970
|
7.467
|
NM_006714
|
SMPDL3A
|
sphingomyelin phosphodiesterase, acid-like 3A
|
|
chr2_+_45878912
|
7.456
|
NM_005400
|
PRKCE
|
protein kinase C, epsilon
|
|
chr1_-_54518790
|
7.399
|
|
TMEM59
|
transmembrane protein 59
|
|
chr17_+_4901199
|
7.337
|
NM_006612
|
KIF1C
|
kinesin family member 1C
|
|
chr5_+_140749830
|
7.322
|
NM_018924
NM_032097
|
PCDHGB3
|
protocadherin gamma subfamily B, 3
|
|
chr10_-_123356158
|
7.239
|
NM_001144915
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr1_-_54518886
|
7.228
|
|
TMEM59
|
transmembrane protein 59
|
|
chr19_+_16254454
|
7.225
|
|
HSH2D
|
hematopoietic SH2 domain containing
|
|
chr2_+_102803432
|
7.188
|
NM_003854
|
IL1RL2
|
interleukin 1 receptor-like 2
|
|
chr3_+_9944511
|
7.075
|
NM_001193380
NM_153480
|
IL17RE
|
interleukin 17 receptor E
|
|
chr12_-_12419702
|
7.073
|
NM_002336
|
LRP6
|
low density lipoprotein receptor-related protein 6
|
|
chr1_-_54518915
|
7.072
|
|
TMEM59
|
transmembrane protein 59
|
|
chr16_+_1290677
|
7.056
|
NM_003294
|
TPSB2
TPSAB1
|
tryptase beta 2 (gene/pseudogene)
tryptase alpha/beta 1
|
|
chr16_+_4838397
|
7.054
|
NM_001253790
NM_001253791
|
LOC440335
|
uncharacterized LOC440335
|
|
chr8_-_33457491
|
7.043
|
|
DUSP26
|
dual specificity phosphatase 26 (putative)
|
|
chr17_-_58603291
|
6.966
|
|
APPBP2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2
|
|
chr1_-_3447974
|
6.959
|
|
MEGF6
|
multiple EGF-like-domains 6
|
|
chr19_-_51456159
|
6.954
|
NM_001077491
NM_001077492
NM_012427
|
KLK5
|
kallikrein-related peptidase 5
|
|
chr1_-_54665670
|
6.929
|
NM_001031672
|
CYB5RL
|
cytochrome b5 reductase-like
|