|
chr20_-_23066828
|
19.557
|
NM_012072
|
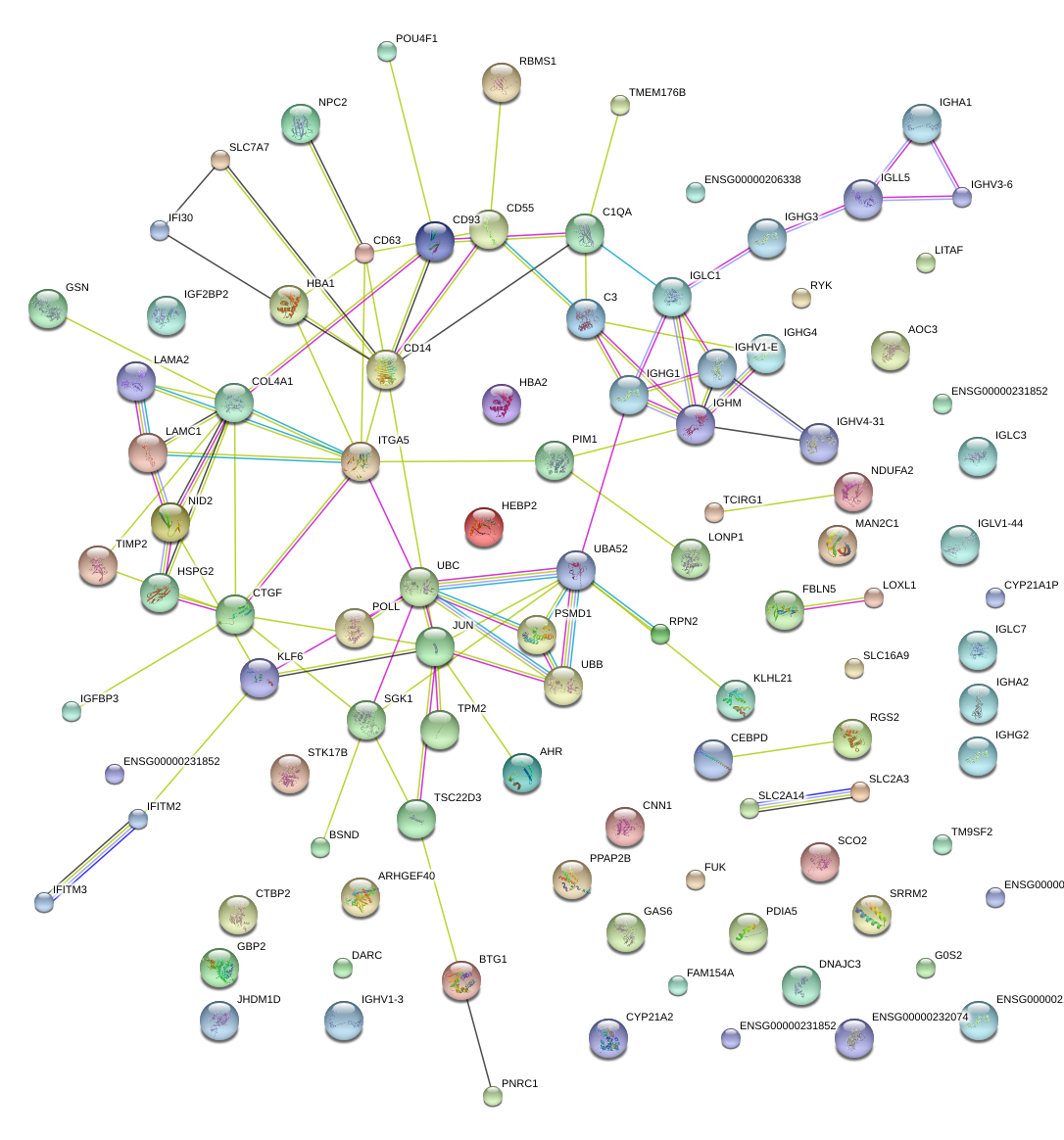
CD93
|
CD93 molecule
|
|
chr6_+_37137882
|
16.830
|
NM_001243186
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr1_+_207495086
|
13.717
|
|
CD55
|
CD55 molecule, decay accelerating factor for complement (Cromer blood group)
|
|
chr8_-_48650683
|
13.421
|
|
CEBPD
|
CCAAT/enhancer binding protein (C/EBP), delta
|
|
chr1_+_207495070
|
12.955
|
|
CD55
|
CD55 molecule, decay accelerating factor for complement (Cromer blood group)
|
|
chr1_+_207495009
|
12.235
|
|
CD55
|
CD55 molecule, decay accelerating factor for complement (Cromer blood group)
|
|
chr1_+_207494816
|
11.805
|
NM_000574
NM_001114752
|
CD55
|
CD55 molecule, decay accelerating factor for complement (Cromer blood group)
|
|
chr10_-_3827205
|
11.110
|
|
KLF6
|
Kruppel-like factor 6
|
|
chr3_+_122785855
|
10.099
|
NM_006810
|
PDIA5
|
protein disulfide isomerase family A, member 5
|
|
chr22_+_23229959
|
9.945
|
NM_001178126
|
IGLL5
IGLC1
|
immunoglobulin lambda-like polypeptide 5
immunoglobulin lambda constant 1 (Mcg marker)
|
|
chr11_-_320925
|
9.888
|
|
IFITM3
|
interferon induced transmembrane protein 3
|
|
chrX_-_106960028
|
9.582
|
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr10_-_126849010
|
9.273
|
|
CTBP2
|
C-terminal binding protein 2
|
|
chr12_-_92539614
|
9.166
|
NM_001731
|
BTG1
|
B-cell translocation gene 1, anti-proliferative
|
|
chr12_-_54813010
|
9.113
|
NM_002205
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr1_+_209848659
|
9.021
|
NM_015714
|
G0S2
|
G0/G1switch 2
|
|
chr1_+_182992694
|
8.930
|
|
|
|
|
chr1_+_55464605
|
8.892
|
NM_057176
|
BSND
|
Bartter syndrome, infantile, with sensorineural deafness (Barttin)
|
|
chr5_-_140013034
|
8.814
|
NM_000591
|
CD14
|
CD14 molecule
|
|
chr1_-_22263676
|
8.522
|
NM_005529
|
HSPG2
|
heparan sulfate proteoglycan 2
|
|
chr11_+_67810446
|
8.490
|
NM_006053
|
TCIRG1
|
T-cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 subunit A3
|
|
chr1_+_22963097
|
8.474
|
NM_015991
|
C1QA
|
complement component 1, q subcomponent, A chain
|
|
chr1_+_192778168
|
8.469
|
NM_002923
|
RGS2
|
regulator of G-protein signaling 2, 24kDa
|
|
chr14_-_52535737
|
7.763
|
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr12_-_54812936
|
7.676
|
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr16_+_2802708
|
7.425
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr3_-_133969563
|
7.378
|
NM_001005861
NM_002958
|
RYK
|
RYK receptor-like tyrosine kinase
|
|
chr7_+_17338182
|
7.372
|
NM_001621
|
AHR
|
aryl hydrocarbon receptor
|
|
chr6_-_134495944
|
6.915
|
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr1_+_159175048
|
6.870
|
NM_001122951
|
DARC
|
Duffy blood group, chemokine receptor
|
|
chr16_+_2802683
|
6.852
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr12_-_54813002
|
6.787
|
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr1_-_59249687
|
6.609
|
NM_002228
|
JUN
|
jun proto-oncogene
|
|
chr17_-_76921205
|
6.511
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr10_-_61469322
|
6.479
|
NM_194298
|
LOC100129721
SLC16A9
|
uncharacterized LOC100129721
solute carrier family 16, member 9 (monocarboxylic acid transporter 9)
|
|
chr17_-_76921076
|
6.431
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr6_-_134496006
|
6.431
|
NM_005627
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr19_+_11649578
|
6.419
|
NM_001299
|
CNN1
|
calponin 1, basic, smooth muscle
|
|
chr12_-_8043743
|
6.286
|
|
SLC2A14
|
solute carrier family 2 (facilitated glucose transporter), member 14
|
|
chr6_+_129204285
|
6.246
|
NM_000426
NM_001079823
|
LAMA2
|
laminin, alpha 2
|
|
chr10_-_126849066
|
6.213
|
NM_001329
|
CTBP2
|
C-terminal binding protein 2
|
|
chr15_-_75660944
|
6.207
|
|
MAN2C1
|
mannosidase, alpha, class 2C, member 1
|
|
chr9_+_124030413
|
6.185
|
|
GSN
|
gelsolin
|
|
chr14_-_23284617
|
6.138
|
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7
|
|
chr1_-_57044980
|
6.076
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr13_-_110959441
|
6.044
|
NM_001845
|
COL4A1
|
collagen, type IV, alpha 1
|
|
chr16_-_11680756
|
6.039
|
NM_001136472
NM_001136473
|
LITAF
|
lipopolysaccharide-induced TNF factor
|
|
chr14_-_107170400
|
5.930
|
|
IGHV4-31
IGHV1-69
IGHA1
IGHG1
IGHG3
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy variable 1-69
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
|
|
chr15_+_74218962
|
5.892
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chrX_-_106959597
|
5.675
|
NM_001015881
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr12_-_56122840
|
5.616
|
NM_001040034
NM_001780
|
CD63
|
CD63 molecule
|
|
chr17_+_41003184
|
5.591
|
NM_003734
|
AOC3
|
amine oxidase, copper containing 3 (vascular adhesion protein 1)
|
|
chr14_-_92414004
|
5.579
|
NM_006329
|
FBLN5
|
fibulin 5
|
|
chr22_-_50964573
|
5.539
|
NM_001169110
|
SCO2
|
SCO cytochrome oxidase deficient homolog 2 (yeast)
|
|
chr19_+_18682613
|
5.526
|
NM_001033930
NM_003333
|
UBA52
|
ubiquitin A-52 residue ribosomal protein fusion product 1
|
|
chr1_-_6662681
|
5.515
|
|
KLHL21
|
kelch-like 21 (Drosophila)
|
|
chr14_+_21538339
|
5.495
|
NM_018071
|
ARHGEF40
|
Rho guanine nucleotide exchange factor (GEF) 40
|
|
chr19_+_18284631
|
5.486
|
|
IFI30
|
interferon, gamma-inducible protein 30
|
|
chr9_+_124030455
|
5.469
|
|
GSN
|
gelsolin
|
|
chr6_+_138725458
|
5.418
|
|
HEBP2
|
heme binding protein 2
|
|
chr12_+_64237381
|
5.394
|
|
|
|
|
chr2_-_161349874
|
5.385
|
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr2_-_197036262
|
5.361
|
|
STK17B
|
serine/threonine kinase 17b
|
|
chr1_+_182992329
|
5.356
|
NM_002293
|
LAMC1
|
laminin, gamma 1 (formerly LAMB2)
|
|
chr22_+_22735134
|
5.310
|
|
CYAT1
|
immunoglobulin lambda light chain-like
|
|
chr13_+_96329392
|
5.309
|
NM_006260
|
DNAJC3
|
DnaJ (Hsp40) homolog, subfamily C, member 3
|
|
chr7_-_45960738
|
5.295
|
NM_000598
NM_001013398
|
IGFBP3
|
insulin-like growth factor binding protein 3
|
|
chr1_-_89591685
|
5.293
|
NM_004120
|
GBP2
|
guanylate binding protein 2, interferon-inducible
|
|
chr3_-_185542807
|
5.286
|
NM_001007225
NM_006548
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr5_-_140012730
|
5.266
|
|
CD14
|
CD14 molecule
|
|
chr14_-_23284565
|
5.236
|
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7
|
|
chr6_+_32006092
|
5.194
|
NM_000500
NM_001128590
|
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2
|
|
chr15_-_75660905
|
5.174
|
NM_006715
|
MAN2C1
|
mannosidase, alpha, class 2C, member 1
|
|
chr7_-_150497398
|
5.172
|
NM_001101311
NM_001101312
NM_001101314
|
TMEM176B
|
transmembrane protein 176B
|
|
chr6_+_31973358
|
5.157
|
NM_000500
NM_001128590
|
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2
|
|
chr1_+_209848994
|
5.144
|
|
G0S2
|
G0/G1switch 2
|
|
chr14_-_106774311
|
5.127
|
|
IGHA1
|
immunoglobulin heavy constant alpha 1
|
|
chr9_-_35689799
|
5.115
|
|
TPM2
|
tropomyosin 2 (beta)
|
|
chr2_-_161350304
|
5.100
|
NM_002897
NM_016836
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr19_-_6720585
|
5.072
|
NM_000064
|
C3
|
complement component 3
|
|
chr7_-_139876718
|
5.045
|
NM_030647
|
JHDM1D
|
jumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae)
|
|
chr13_+_100153771
|
5.038
|
|
TM9SF2
|
transmembrane 9 superfamily member 2
|
|
chr14_-_74960003
|
4.960
|
|
NPC2
|
Niemann-Pick disease, type C2
|
|
chr14_-_74960035
|
4.933
|
NM_006432
|
NPC2
|
Niemann-Pick disease, type C2
|
|
chr10_-_103347882
|
4.930
|
|
POLL
|
polymerase (DNA directed), lambda
|
|
chr17_-_76921453
|
4.881
|
NM_003255
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr12_-_56122800
|
4.853
|
|
CD63
|
CD63 molecule
|
|
chr13_-_79177682
|
4.841
|
NM_006237
|
POU4F1
|
POU class 4 homeobox 1
|
|
chr16_+_222845
|
4.835
|
NM_000517
|
HBA1
HBA2
|
hemoglobin, alpha 1
hemoglobin, alpha 2
|
|
chr14_-_106092256
|
4.828
|
|
IGHG4
|
immunoglobulin heavy constant gamma 4 (G4m marker)
|
|
chr20_+_35807695
|
4.811
|
|
RPN2
|
ribophorin II
|
|
chr6_+_89791510
|
4.788
|
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr20_+_35807430
|
4.762
|
NM_001135771
NM_002951
|
RPN2
|
ribophorin II
|
|
chr14_-_92413738
|
4.749
|
|
FBLN5
|
fibulin 5
|
|
chr16_+_70502824
|
4.737
|
|
FUK
|
fucokinase
|
|
chr6_-_132272311
|
4.713
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chr20_+_35807747
|
4.700
|
|
RPN2
|
ribophorin II
|
|
chr20_+_35807522
|
4.690
|
|
RPN2
|
ribophorin II
|
|
chr13_-_114567040
|
4.675
|
NM_000820
|
GAS6
|
growth arrest-specific 6
|
|
chr14_-_74959976
|
4.666
|
|
NPC2
|
Niemann-Pick disease, type C2
|
|
chr14_-_52535711
|
4.649
|
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr1_-_57045129
|
4.642
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr5_-_39425297
|
4.631
|
NM_001244871
NM_001343
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr10_-_105212149
|
4.626
|
NM_015916
|
CALHM2
|
calcium homeostasis modulator 2
|
|
chr21_-_28217272
|
4.613
|
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr14_-_106725620
|
4.600
|
|
IGHV4-31
IGHV3-23
IGHA1
IGHG1
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy variable 3-23
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant mu
|
|
chr17_+_41006094
|
4.598
|
|
AOC3
|
amine oxidase, copper containing 3 (vascular adhesion protein 1)
|
|
chr16_-_31106049
|
4.575
|
|
VKORC1
|
vitamin K epoxide reductase complex, subunit 1
|
|
chr22_+_22786283
|
4.573
|
|
|
|
|
chr3_-_185542744
|
4.560
|
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr15_+_74218786
|
4.549
|
NM_005576
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr17_+_55055467
|
4.513
|
NM_021626
|
SCPEP1
|
serine carboxypeptidase 1
|
|
chr16_-_15835382
|
4.512
|
|
MYH11
|
myosin, heavy chain 11, smooth muscle
|
|
chr17_-_66453561
|
4.508
|
NM_017983
|
WIPI1
|
WD repeat domain, phosphoinositide interacting 1
|
|
chr15_+_74218811
|
4.497
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr8_+_97506020
|
4.488
|
|
SDC2
|
syndecan 2
|
|
chr3_+_5021141
|
4.488
|
|
BHLHE40
|
basic helix-loop-helix family, member e40
|
|
chr13_-_114566951
|
4.430
|
|
GAS6
|
growth arrest-specific 6
|
|
chr2_+_17935124
|
4.407
|
NM_182625
|
GEN1
|
Gen endonuclease homolog 1 (Drosophila)
|
|
chr6_-_144385647
|
4.403
|
NM_001080951
NM_001080955
|
PLAGL1
|
pleiomorphic adenoma gene-like 1
|
|
chr11_+_117049838
|
4.371
|
NM_001040455
|
SIDT2
|
SID1 transmembrane family, member 2
|
|
chr21_-_28217692
|
4.369
|
NM_006988
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr19_-_54746599
|
4.338
|
NM_024318
|
LILRB3
LILRA6
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 6
|
|
chr11_+_67806454
|
4.336
|
NM_006019
|
TCIRG1
|
T-cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 subunit A3
|
|
chr14_+_101193313
|
4.325
|
|
DLK1
|
delta-like 1 homolog (Drosophila)
|
|
chr3_-_50378238
|
4.315
|
NM_001206957
NM_007182
NM_170714
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1
|
|
chr1_-_178062734
|
4.302
|
|
LOC100302401
|
uncharacterized LOC100302401
|
|
chrX_-_38079980
|
4.292
|
NM_001170750
NM_001170751
NM_001170752
NM_006307
|
SRPX
|
sushi-repeat containing protein, X-linked
|
|
chr1_+_12227050
|
4.287
|
NM_001066
|
TNFRSF1B
|
tumor necrosis factor receptor superfamily, member 1B
|
|
chr10_-_103347978
|
4.279
|
NM_001174084
NM_001174085
NM_013274
|
POLL
|
polymerase (DNA directed), lambda
|
|
chr10_-_118429469
|
4.275
|
NM_144661
|
C10orf82
|
chromosome 10 open reading frame 82
|
|
chr11_+_118401873
|
4.258
|
|
TMEM25
|
transmembrane protein 25
|
|
chr17_-_8055684
|
4.255
|
NM_002616
|
PER1
|
period homolog 1 (Drosophila)
|
|
chr22_+_45898690
|
4.233
|
NM_001996
NM_006485
NM_006486
NM_006487
|
FBLN1
|
fibulin 1
|
|
chr14_-_52535945
|
4.222
|
NM_007361
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr9_+_90341030
|
4.210
|
|
CTSL1
|
cathepsin L1
|
|
chr1_+_180123906
|
4.195
|
NM_001004128
NM_002826
|
QSOX1
|
quiescin Q6 sulfhydryl oxidase 1
|
|
chr6_-_144329342
|
4.188
|
NM_001080952
NM_001080953
NM_001080954
NM_001080956
NM_002656
NM_006718
|
PLAGL1
HYMAI
|
pleiomorphic adenoma gene-like 1
hydatidiform mole associated and imprinted (non-protein coding)
|
|
chr15_-_48470453
|
4.177
|
NM_016132
|
MYEF2
|
myelin expression factor 2
|
|
chr9_+_90340973
|
4.168
|
NM_001912
NM_145918
|
CTSL1
|
cathepsin L1
|
|
chr14_-_107078578
|
4.160
|
|
|
|
|
chr16_+_29831714
|
4.149
|
NM_005115
NM_017458
|
MVP
|
major vault protein
|
|
chr9_+_90341022
|
4.145
|
|
CTSL1
|
cathepsin L1
|
|
chr7_+_100199881
|
4.142
|
NM_002593
|
PCOLCE
|
procollagen C-endopeptidase enhancer
|
|
chr13_-_114567011
|
4.138
|
|
GAS6
|
growth arrest-specific 6
|
|
chr1_+_180124031
|
4.138
|
|
QSOX1
|
quiescin Q6 sulfhydryl oxidase 1
|
|
chr1_+_180124023
|
4.127
|
|
QSOX1
|
quiescin Q6 sulfhydryl oxidase 1
|
|
chr13_+_110959592
|
4.123
|
NM_001846
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr14_+_101193275
|
4.116
|
|
DLK1
|
delta-like 1 homolog (Drosophila)
|
|
chr14_-_94856926
|
4.102
|
NM_001002235
NM_001002236
NM_001127700
NM_001127701
NM_001127702
NM_001127703
NM_001127704
NM_001127705
NM_001127706
NM_001127707
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr19_-_11689765
|
4.087
|
NM_001111035
NM_001111036
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr16_-_31106125
|
4.074
|
NM_024006
NM_206824
|
VKORC1
|
vitamin K epoxide reductase complex, subunit 1
|
|
chr20_+_19870209
|
4.071
|
NM_018993
|
RIN2
|
Ras and Rab interactor 2
|
|
chr1_-_120612301
|
4.062
|
NM_001200001
NM_024408
|
NOTCH2
|
notch 2
|
|
chr8_+_90770201
|
4.018
|
|
RIPK2
|
receptor-interacting serine-threonine kinase 2
|
|
chr1_+_145209067
|
4.007
|
NM_203458
|
NOTCH2NL
|
notch 2 N-terminal like
|
|
chr8_+_97505895
|
4.006
|
|
SDC2
|
syndecan 2
|
|
chr14_-_106641585
|
3.991
|
|
IGHV4-31
IGHA1
IGHA2
IGHG1
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr9_-_35689902
|
3.978
|
NM_003289
NM_213674
|
TPM2
|
tropomyosin 2 (beta)
|
|
chr14_-_106110894
|
3.958
|
|
IGHG2
|
immunoglobulin heavy constant gamma 2 (G2m marker)
|
|
chr15_+_74218794
|
3.949
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr11_+_67806517
|
3.909
|
|
TCIRG1
|
T-cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 subunit A3
|
|
chr11_-_67120943
|
3.896
|
NM_021173
|
POLD4
|
polymerase (DNA-directed), delta 4
|
|
chr14_-_100070442
|
3.875
|
NM_001144995
|
CCDC85C
|
coiled-coil domain containing 85C
|
|
chr7_+_102553343
|
3.874
|
NM_001031692
NM_005824
|
LRRC17
|
leucine rich repeat containing 17
|
|
chr8_+_144640476
|
3.870
|
NM_024736
|
GSDMD
|
gasdermin D
|
|
chr2_-_209119751
|
3.861
|
NM_005896
|
IDH1
|
isocitrate dehydrogenase 1 (NADP+), soluble
|
|
chr2_+_10183630
|
3.851
|
NM_003597
NM_001177716
|
KLF11
|
Kruppel-like factor 11
|
|
chr16_-_88923233
|
3.850
|
|
GALNS
|
galactosamine (N-acetyl)-6-sulfate sulfatase
|
|
chr9_+_124030370
|
3.839
|
NM_001127662
NM_001127663
NM_001127664
NM_001127665
NM_001127666
NM_001127667
NM_198252
|
GSN
|
gelsolin
|
|
chr16_-_46865039
|
3.829
|
NM_001001436
|
C16orf87
|
chromosome 16 open reading frame 87
|
|
chr1_-_113249677
|
3.822
|
NM_001042679
|
RHOC
|
ras homolog gene family, member C
|
|
chr11_+_842821
|
3.814
|
NM_001025238
NM_001025239
NM_003271
NM_001025235
NM_001025236
|
TSPAN4
|
tetraspanin 4
|
|
chr14_+_101193190
|
3.812
|
NM_003836
|
DLK1
|
delta-like 1 homolog (Drosophila)
|
|
chr18_+_21594349
|
3.811
|
NM_001135993
NM_001243425
|
TTC39C
|
tetratricopeptide repeat domain 39C
|
|
chr22_-_50964856
|
3.798
|
NM_001169109
|
SCO2
|
SCO cytochrome oxidase deficient homolog 2 (yeast)
|
|
chr9_+_90341123
|
3.791
|
|
CTSL1
|
cathepsin L1
|
|
chr1_-_57045214
|
3.791
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr13_+_96329409
|
3.788
|
|
DNAJC3
|
DnaJ (Hsp40) homolog, subfamily C, member 3
|
|
chr2_-_220436010
|
3.783
|
|
OBSL1
|
obscurin-like 1
|
|
chr14_-_23285011
|
3.779
|
NM_001126105
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7
|
|
chr19_+_18284578
|
3.778
|
NM_006332
|
IFI30
|
interferon, gamma-inducible protein 30
|
|
chr1_-_115053677
|
3.757
|
|
TRIM33
|
tripartite motif containing 33
|
|
chr1_+_180124013
|
3.753
|
|
QSOX1
|
quiescin Q6 sulfhydryl oxidase 1
|
|
chr4_-_2263808
|
3.732
|
|
MXD4
|
MAX dimerization protein 4
|
|
chr6_+_72596648
|
3.726
|
NM_014989
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr1_-_113250022
|
3.723
|
NM_001042678
NM_175744
|
RHOC
|
ras homolog gene family, member C
|
|
chr16_-_31106093
|
3.710
|
|
VKORC1
|
vitamin K epoxide reductase complex, subunit 1
|
|
chr13_+_96329398
|
3.691
|
|
DNAJC3
|
DnaJ (Hsp40) homolog, subfamily C, member 3
|
|
chr3_+_122296432
|
3.662
|
NM_001113523
|
PARP15
|
poly (ADP-ribose) polymerase family, member 15
|
|
chr11_+_113930430
|
3.657
|
NM_006006
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr8_+_56792399
|
3.646
|
|
LYN
|
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog
|
|
chr19_+_39899749
|
3.643
|
|
|
|
|
chr11_-_615649
|
3.633
|
NM_004031
|
IRF7
|
interferon regulatory factor 7
|
|
chr14_-_107013474
|
3.633
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr15_+_74466887
|
3.625
|
NM_201526
|
ISLR
|
immunoglobulin superfamily containing leucine-rich repeat
|
|
chr3_+_148709252
|
3.616
|
|
GYG1
|
glycogenin 1
|
|
chr1_-_120612019
|
3.615
|
|
NOTCH2
|
notch 2
|
|
chr8_-_28243589
|
3.611
|
|
ZNF395
|
zinc finger protein 395
|
|
chr2_-_111230651
|
3.605
|
NM_001205288
NM_033514
|
LIMS3-LOC440895
LIMS3L
LIMS3
|
LIMS3-LOC440895 readthrough
LIM and senescent cell antigen-like domains 3-like
LIM and senescent cell antigen-like domains 3
|