|
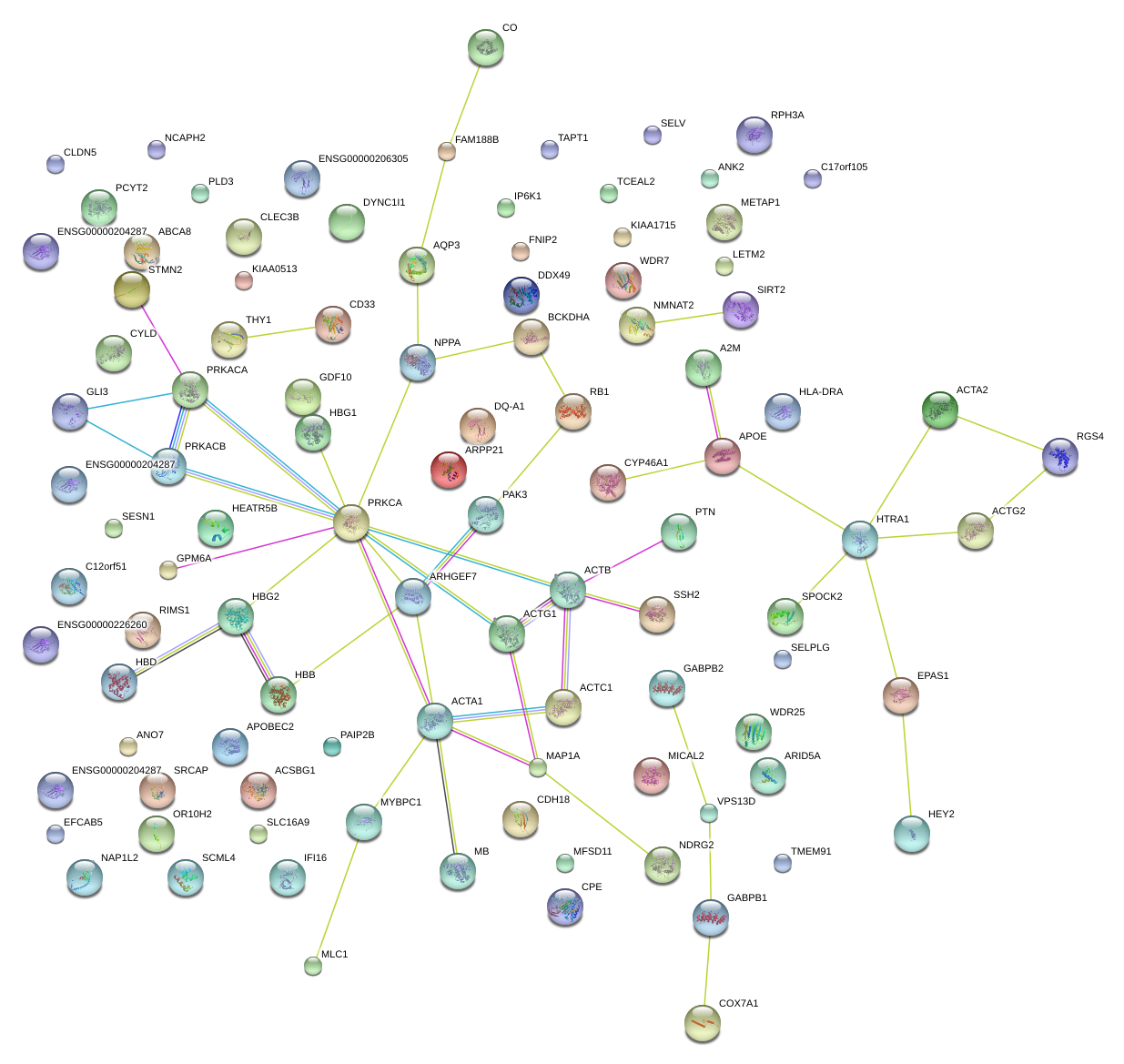
chr11_-_5248180
|
82.041
|
NM_000518
|
HBB
|
hemoglobin, beta
|
|
chr8_+_80523048
|
65.290
|
NM_001199214
NM_007029
|
STMN2
|
stathmin-like 2
|
|
chr12_-_9268513
|
59.342
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr19_+_45409003
|
47.019
|
NM_000041
|
APOE
|
apolipoprotein E
|
|
chr3_+_45067750
|
42.103
|
NM_003278
|
CLEC3B
|
C-type lectin domain family 3, member B
|
|
chr12_-_9268475
|
41.329
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr19_-_39390168
|
39.464
|
NM_012237
NM_001193286
NM_030593
|
SIRT2
|
sirtuin 2
|
|
chr12_-_9268522
|
39.133
|
NM_000014
|
A2M
|
alpha-2-macroglobulin
|
|
chr4_+_113739203
|
38.873
|
NM_001127493
|
ANK2
|
ankyrin 2, neuronal
|
|
chr6_+_32407618
|
38.757
|
NM_019111
|
HLA-DRA
|
major histocompatibility complex, class II, DR alpha
|
|
chr6_-_108145507
|
37.937
|
NM_198081
|
SCML4
|
sex comb on midleg-like 4 (Drosophila)
|
|
chr1_-_11907741
|
37.835
|
NM_006172
|
NPPA
|
natriuretic peptide A
|
|
chr14_+_100150596
|
37.816
|
NM_006668
|
CYP46A1
|
cytochrome P450, family 46, subfamily A, polypeptide 1
|
|
chr12_-_9268440
|
37.686
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr17_+_41857802
|
37.132
|
NM_001136483
|
C17orf105
|
chromosome 17 open reading frame 105
|
|
chr10_-_73848263
|
36.306
|
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2
|
|
chr7_+_30951414
|
35.752
|
NM_198098
|
AQP1
|
aquaporin 1 (Colton blood group)
|
|
chr16_+_30710461
|
34.827
|
NM_006662
|
SRCAP
|
Snf2-related CREBBP activator protein
|
|
chr3_+_35722428
|
34.583
|
NM_198399
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr7_-_137028533
|
33.644
|
NM_002825
|
PTN
|
pleiotrophin
|
|
chr14_-_21493122
|
33.260
|
NM_201538
NM_201539
NM_201540
NM_201541
|
NDRG2
|
NDRG family member 2
|
|
chr18_+_54318561
|
32.691
|
NM_015285
NM_052834
|
WDR7
|
WD repeat domain 7
|
|
chr19_-_36643770
|
32.327
|
NM_001864
|
COX7A1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle)
|
|
chr9_-_33447527
|
32.218
|
NM_004925
|
AQP3
|
aquaporin 3 (Gill blood group)
|
|
chr4_-_176733901
|
31.535
|
|
GPM6A
|
glycoprotein M6A
|
|
chr4_-_176923572
|
31.421
|
NM_005277
NM_201592
|
GPM6A
|
glycoprotein M6A
|
|
chr7_+_95401817
|
30.989
|
NM_001135556
NM_001135557
NM_004411
|
DYNC1I1
|
dynein, cytoplasmic 1, intermediate chain 1
|
|
chr1_+_163038395
|
30.613
|
NM_001102445
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr12_+_101988746
|
30.505
|
NM_002465
NM_206819
NM_206820
NM_206821
|
MYBPC1
|
myosin binding protein C, slow type
|
|
chr22_-_36013303
|
30.156
|
NM_005368
NM_203378
|
MB
|
myoglobin
|
|
chr22_-_50523737
|
29.859
|
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1
|
|
chrX_+_101380659
|
29.686
|
NM_080390
|
TCEAL2
|
transcription elongation factor A (SII)-like 2
|
|
chr11_-_5271035
|
29.621
|
NM_000559
|
HBG1
|
hemoglobin, gamma A
|
|
chr17_-_28257017
|
29.496
|
NM_033389
|
SSH2
|
slingshot homolog 2 (Drosophila)
|
|
chr3_-_49823953
|
29.353
|
|
IP6K1
|
inositol hexakisphosphate kinase 1
|
|
chr5_-_19988293
|
29.211
|
NM_001167667
NM_004934
|
CDH18
|
cadherin 18, type 2
|
|
chr8_+_80523304
|
29.192
|
|
STMN2
|
stathmin-like 2
|
|
chr4_+_166299942
|
28.938
|
NM_001873
|
CPE
|
carboxypeptidase E
|
|
chr22_-_19511777
|
28.886
|
|
CLDN5
|
claudin 5
|
|
chr12_+_113229548
|
28.590
|
NM_001143854
NM_014954
|
RPH3A
|
rabphilin 3A homolog (mouse)
|
|
chr22_+_50946627
|
27.684
|
NM_001185011
NM_014551
NM_152299
|
NCAPH2
|
non-SMC condensin II complex, subunit H2
|
|
chr4_-_176733413
|
27.509
|
|
GPM6A
|
glycoprotein M6A
|
|
chr2_-_71454200
|
27.469
|
NM_020459
|
PAIP2B
|
poly(A) binding protein interacting protein 2B
|
|
chrX_+_110187468
|
27.227
|
NM_001128166
NM_001128167
|
PAK3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr17_+_28256873
|
26.854
|
NM_001145053
|
EFCAB5
|
EF-hand calcium binding domain 5
|
|
chr17_+_74734106
|
26.747
|
|
MFSD11
|
major facilitator superfamily domain containing 11
|
|
chr22_-_50523780
|
26.741
|
NM_139202
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1
|
|
chr22_-_50523853
|
26.656
|
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1
|
|
chr11_-_119294242
|
26.634
|
NM_006288
|
THY1
|
Thy-1 cell surface antigen
|
|
chr12_-_112819755
|
26.345
|
NM_001109662
|
C12orf51
|
chromosome 12 open reading frame 51
|
|
chr6_+_126070813
|
26.093
|
|
HEY2
|
hairy/enhancer-of-split related with YRPW motif 2
|
|
chr22_-_19512001
|
26.064
|
|
CLDN5
|
claudin 5
|
|
chr22_-_50524357
|
25.147
|
NM_015166
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1
|
|
chr2_+_46524901
|
24.493
|
|
EPAS1
|
endothelial PAS domain protein 1
|
|
chr2_+_242127923
|
24.434
|
NM_001001666
NM_001001891
|
ANO7
|
anoctamin 7
|
|
chr19_+_19030523
|
24.371
|
|
DDX49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49
|
|
chr14_+_100842728
|
24.348
|
NM_001161476
NM_024515
|
WDR25
|
WD repeat domain 25
|
|
chr6_+_41020939
|
24.344
|
NM_006789
|
APOBEC2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2
|
|
chr2_-_176866925
|
24.077
|
NM_030650
|
KIAA1715
|
KIAA1715
|
|
chr2_+_46524448
|
24.034
|
NM_001430
|
EPAS1
|
endothelial PAS domain protein 1
|
|
chrX_-_72434656
|
23.859
|
|
NAP1L2
|
nucleosome assembly protein 1-like 2
|
|
chr1_+_151043079
|
23.838
|
NM_144618
|
GABPB2
|
GA binding protein transcription factor, beta subunit 2
|
|
chr6_+_73076431
|
23.807
|
NM_001168411
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr1_-_183274008
|
23.756
|
NM_170706
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2
|
|
chr19_+_40854529
|
23.618
|
|
PLD3
|
phospholipase D family, member 3
|
|
chr19_+_19030491
|
23.382
|
|
DDX49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49
|
|
chr10_-_61469322
|
23.300
|
NM_194298
|
LOC100129721
SLC16A9
|
uncharacterized LOC100129721
solute carrier family 16, member 9 (monocarboxylic acid transporter 9)
|
|
chr4_-_16228071
|
22.969
|
|
TAPT1
|
transmembrane anterior posterior transformation 1
|
|
chr3_-_49823972
|
22.312
|
NM_001006115
NM_153273
|
IP6K1
|
inositol hexakisphosphate kinase 1
|
|
chr2_-_37311458
|
22.200
|
NM_019024
|
HEATR5B
|
HEAT repeat containing 5B
|
|
chr10_+_124220990
|
22.088
|
NM_002775
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr13_+_48877950
|
22.079
|
|
RB1
|
retinoblastoma 1
|
|
chr4_+_159690208
|
21.777
|
|
FNIP2
|
folliculin interacting protein 2
|
|
chr4_+_100871625
|
21.721
|
|
LOC256880
|
uncharacterized LOC256880
|
|
chr12_-_109027565
|
21.684
|
|
SELPLG
|
selectin P ligand
|
|
chr8_+_38243958
|
21.504
|
NM_001199659
NM_144652
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2
|
|
chr6_-_109415514
|
21.440
|
NM_014454
|
SESN1
|
sestrin 1
|
|
chr15_+_43809805
|
21.424
|
NM_002373
|
MAP1A
|
microtubule-associated protein 1A
|
|
chr10_-_90750955
|
21.383
|
NM_001141945
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr19_+_51728369
|
21.314
|
|
CD33
|
CD33 molecule
|
|
chr16_+_85061303
|
21.150
|
NM_014732
|
KIAA0513
|
KIAA0513
|
|
chr2_+_97202463
|
20.987
|
NM_212481
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like)
|
|
chr19_+_41903718
|
20.795
|
|
BCKDHA
|
branched chain keto acid dehydrogenase E1, alpha polypeptide
|
|
chr17_-_79869033
|
20.770
|
|
PCYT2
|
phosphate cytidylyltransferase 2, ethanolamine
|
|
chr11_+_12132063
|
20.638
|
NM_014632
|
MICAL2
|
microtubule associated monoxygenase, calponin and LIM domain containing 2
|
|
chr19_-_14228560
|
20.585
|
|
PRKACA
|
protein kinase, cAMP-dependent, catalytic, alpha
|
|
chr7_-_42276617
|
20.512
|
NM_000168
|
GLI3
|
GLI family zinc finger 3
|
|
chr17_-_66951381
|
20.400
|
NM_007168
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8
|
|
chr10_-_48438975
|
20.309
|
NM_004962
|
GDF10
|
growth differentiation factor 10
|
|
chr16_+_50776012
|
20.153
|
NM_001042412
|
CYLD
|
cylindromatosis (turban tumor syndrome)
|
|
chr10_-_69597768
|
20.150
|
NM_021800
NM_201262
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12
|
|
chr14_-_24584125
|
20.115
|
|
NRL
|
neural retina leucine zipper
|
|
chr1_-_54872075
|
20.101
|
NM_001009955
|
SSBP3
|
single stranded DNA binding protein 3
|
|
chr16_+_71392615
|
19.837
|
NM_001740
NM_007088
|
CALB2
|
calbindin 2
|
|
chr16_+_50775960
|
19.743
|
NM_001042355
NM_015247
|
CYLD
|
cylindromatosis (turban tumor syndrome)
|
|
chr6_-_13487772
|
19.733
|
NM_001242629
NM_018988
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1
|
|
chr10_+_86004808
|
19.706
|
NM_001012720
NM_001012722
NM_002921
|
RGR
|
retinal G protein coupled receptor
|
|
chr2_-_87017948
|
19.646
|
|
CD8A
|
CD8a molecule
|
|
chr6_-_41863086
|
19.626
|
NM_018561
|
USP49
|
ubiquitin specific peptidase 49
|
|
chr20_+_1246959
|
19.612
|
NM_014723
|
SNPH
|
syntaphilin
|
|
chr3_-_127541975
|
19.498
|
NM_001003794
|
MGLL
|
monoglyceride lipase
|
|
chr1_-_160681552
|
19.322
|
NM_001778
|
CD48
|
CD48 molecule
|
|
chr1_-_20141593
|
19.253
|
NM_019062
|
RNF186
|
ring finger protein 186
|
|
chr18_-_47807676
|
19.119
|
|
MBD1
|
methyl-CpG binding domain protein 1
|
|
chr16_+_71392637
|
19.063
|
|
CALB2
|
calbindin 2
|
|
chr19_-_14952688
|
18.971
|
NM_001005190
|
OR7A10
|
olfactory receptor, family 7, subfamily A, member 10
|
|
chr6_-_41130864
|
18.970
|
|
TREM2
|
triggering receptor expressed on myeloid cells 2
|
|
chr16_-_30582951
|
18.812
|
|
ZNF688
|
zinc finger protein 688
|
|
chr4_+_88896868
|
18.754
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr5_+_157170702
|
18.750
|
NM_173491
|
LSM11
|
LSM11, U7 small nuclear RNA associated
|
|
chr19_+_51728319
|
18.743
|
NM_001082618
NM_001177608
NM_001772
|
CD33
|
CD33 molecule
|
|
chr18_-_52969714
|
18.698
|
NM_001243236
|
TCF4
|
transcription factor 4
|
|
chr4_+_128886539
|
18.668
|
|
C4orf29
|
chromosome 4 open reading frame 29
|
|
chr2_+_64751545
|
18.652
|
|
AFTPH
|
aftiphilin
|
|
chr2_-_201936386
|
18.626
|
|
FAM126B
|
family with sequence similarity 126, member B
|
|
chr2_-_211179765
|
18.623
|
NM_079420
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast
|
|
chr4_+_88896899
|
18.608
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr1_+_163038564
|
18.599
|
NM_005613
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr7_+_97361270
|
18.598
|
NM_003182
NM_013996
NM_013997
NM_013998
|
TAC1
|
tachykinin, precursor 1
|
|
chr6_+_99282579
|
18.464
|
NM_005604
|
POU3F2
|
POU class 3 homeobox 2
|
|
chr19_+_4969123
|
18.385
|
NM_015015
|
KDM4B
|
lysine (K)-specific demethylase 4B
|
|
chr10_-_99393851
|
18.367
|
NM_001098831
|
MORN4
|
MORN repeat containing 4
|
|
chr14_-_73360792
|
18.313
|
NM_012074
|
DPF3
|
D4, zinc and double PHD fingers, family 3
|
|
chr2_+_233925104
|
18.305
|
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa
|
|
chr13_+_28494136
|
18.257
|
NM_000209
|
PDX1
|
pancreatic and duodenal homeobox 1
|
|
chr6_-_84418705
|
18.240
|
NM_001242792
|
SNAP91
|
synaptosomal-associated protein, 91kDa homolog (mouse)
|
|
chr12_+_6898705
|
18.189
|
|
CD4
|
CD4 molecule
|
|
chr1_-_220445747
|
18.082
|
|
RAB3GAP2
|
RAB3 GTPase activating protein subunit 2 (non-catalytic)
|
|
chrX_-_72434623
|
18.049
|
|
NAP1L2
|
nucleosome assembly protein 1-like 2
|
|
chr5_-_24645084
|
18.044
|
NM_006727
|
CDH10
|
cadherin 10, type 2 (T2-cadherin)
|
|
chr12_+_53442732
|
17.989
|
NM_015319
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr6_-_112194430
|
17.936
|
|
FYN
|
FYN oncogene related to SRC, FGR, YES
|
|
chr17_-_26220372
|
17.855
|
NM_001076680
|
C17orf108
|
chromosome 17 open reading frame 108
|
|
chr4_+_113970784
|
17.848
|
NM_001148
NM_020977
|
ANK2
|
ankyrin 2, neuronal
|
|
chr4_-_176923480
|
17.791
|
|
GPM6A
|
glycoprotein M6A
|
|
chr12_-_109027615
|
17.778
|
|
SELPLG
|
selectin P ligand
|
|
chr1_-_155532152
|
17.712
|
|
ASH1L
|
ash1 (absent, small, or homeotic)-like (Drosophila)
|
|
chr19_+_40854331
|
17.642
|
NM_001031696
NM_012268
|
PLD3
|
phospholipase D family, member 3
|
|
chr4_-_176923533
|
17.571
|
|
GPM6A
|
glycoprotein M6A
|
|
chr1_-_54871978
|
17.556
|
NM_018070
NM_145716
|
SSBP3
|
single stranded DNA binding protein 3
|
|
chr16_-_30583727
|
17.532
|
NM_001024683
NM_145271
|
ZNF688
|
zinc finger protein 688
|
|
chr5_+_140868721
|
17.478
|
NM_018929
NM_032407
|
PCDHGC5
|
protocadherin gamma subfamily C, 5
|
|
chr17_-_66287256
|
17.445
|
NM_001174166
NM_004694
|
SLC16A6
|
solute carrier family 16, member 6 (monocarboxylic acid transporter 7)
|
|
chr2_-_1926501
|
17.400
|
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr14_-_106237675
|
17.384
|
|
IGHM
|
immunoglobulin heavy constant mu
|
|
chr17_+_3377346
|
17.290
|
NM_001128085
|
ASPA
|
aspartoacylase
|
|
chr6_-_112194648
|
17.283
|
NM_002037
|
FYN
|
FYN oncogene related to SRC, FGR, YES
|
|
chr4_+_88896801
|
17.258
|
NM_000582
NM_001040058
NM_001040060
NM_001251829
NM_001251830
|
SPP1
|
secreted phosphoprotein 1
|
|
chr1_+_100111430
|
17.150
|
NM_017734
|
PALMD
|
palmdelphin
|
|
chr17_-_56618178
|
17.139
|
NM_001198713
|
SEPT4
|
septin 4
|
|
chr15_+_42840991
|
17.129
|
NM_001130447
NM_018097
|
HAUS2
|
HAUS augmin-like complex, subunit 2
|
|
chr5_-_169724614
|
17.111
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr15_-_42840910
|
17.017
|
|
LRRC57
|
leucine rich repeat containing 57
|
|
chr2_+_233925035
|
16.908
|
NM_001017915
NM_005541
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa
|
|
chr12_+_119616594
|
16.855
|
NM_014365
|
HSPB8
|
heat shock 22kDa protein 8
|
|
chr3_-_52719514
|
16.786
|
NM_018313
|
PBRM1
|
polybromo 1
|
|
chr7_-_152132924
|
16.783
|
|
MLL3
|
myeloid/lymphoid or mixed-lineage leukemia 3
|
|
chr1_-_54871681
|
16.755
|
|
SSBP3
|
single stranded DNA binding protein 3
|
|
chr3_-_128879873
|
16.681
|
NM_001204890
NM_001199469
NM_020701
|
ISY1-RAB43
ISY1
|
ISY1-RAB43 readthrough
ISY1 splicing factor homolog (S. cerevisiae)
|
|
chr3_-_101231973
|
16.468
|
NM_001077203
NM_020654
|
SENP7
|
SUMO1/sentrin specific peptidase 7
|
|
chr19_+_48972574
|
16.460
|
|
CYTH2
|
cytohesin 2
|
|
chr3_+_4535106
|
16.448
|
|
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1
|
|
chr10_+_115310589
|
16.277
|
NM_001177660
|
HABP2
|
hyaluronan binding protein 2
|
|
chr1_+_101702561
|
16.203
|
|
S1PR1
|
sphingosine-1-phosphate receptor 1
|
|
chr17_+_59477473
|
16.201
|
|
TBX2
|
T-box 2
|
|
chr12_-_109027654
|
16.109
|
NM_003006
|
SELPLG
|
selectin P ligand
|
|
chrX_-_48931645
|
16.082
|
NM_007213
|
PRAF2
|
PRA1 domain family, member 2
|
|
chrX_-_72434667
|
16.061
|
NM_021963
|
NAP1L2
|
nucleosome assembly protein 1-like 2
|
|
chr11_+_73661363
|
16.030
|
NM_153614
|
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13
|
|
chr10_+_106034886
|
16.012
|
NM_001191014
NM_001191015
|
GSTO2
|
glutathione S-transferase omega 2
|
|
chr10_-_99393175
|
15.976
|
NM_178832
|
MORN4
|
MORN repeat containing 4
|
|
chr6_-_41130921
|
15.799
|
NM_018965
|
TREM2
|
triggering receptor expressed on myeloid cells 2
|
|
chr16_+_31366519
|
15.745
|
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit)
|
|
chr3_+_31574119
|
15.737
|
|
STT3B
|
STT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae)
|
|
chr5_-_88199868
|
15.622
|
NM_001131005
NM_001193347
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr4_+_159593233
|
15.526
|
NM_004453
|
ETFDH
|
electron-transferring-flavoprotein dehydrogenase
|
|
chr11_-_57004674
|
15.367
|
NM_005161
|
APLNR
|
apelin receptor
|
|
chr4_-_89205667
|
15.359
|
|
PPM1K
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K
|
|
chr1_+_101702304
|
15.355
|
NM_001400
|
S1PR1
|
sphingosine-1-phosphate receptor 1
|
|
chr12_-_48152860
|
15.304
|
NM_001098531
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3
|
|
chr2_+_143886898
|
15.252
|
NM_018460
|
ARHGAP15
|
Rho GTPase activating protein 15
|
|
chr3_+_150264514
|
15.220
|
NM_032025
|
EIF2A
|
eukaryotic translation initiation factor 2A, 65kDa
|
|
chr4_+_88896838
|
15.212
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr19_-_14228362
|
15.197
|
NM_002730
|
PRKACA
|
protein kinase, cAMP-dependent, catalytic, alpha
|
|
chr7_+_135347211
|
15.166
|
NM_001130929
|
SLC13A4
C7orf73
|
solute carrier family 13 (sodium/sulfate symporters), member 4
chromosome 7 open reading frame 73
|
|
chr17_+_68165660
|
15.134
|
NM_000891
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2
|
|
chr15_-_23208356
|
15.049
|
|
WHAMMP3
|
WAS protein homolog associated with actin, golgi membranes and microtubules pseudogene 3
|
|
chr12_-_16759939
|
14.988
|
NM_001243613
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr12_+_53497273
|
14.812
|
NM_003578
|
SOAT2
|
sterol O-acyltransferase 2
|
|
chrX_+_153775828
|
14.745
|
|
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma
|
|
chr8_-_67090690
|
14.742
|
NM_000756
|
CRH
|
corticotropin releasing hormone
|
|
chr19_-_38714781
|
14.739
|
NM_001135155
NM_004647
|
DPF1
|
D4, zinc and double PHD fingers family 1
|
|
chr2_-_188312872
|
14.715
|
NM_005795
|
CALCRL
|
calcitonin receptor-like
|
|
chr12_+_108909049
|
14.710
|
NM_007076
|
FICD
|
FIC domain containing
|
|
chr10_-_75255718
|
14.659
|
|
PPP3CB
|
protein phosphatase 3, catalytic subunit, beta isozyme
|
|
chr12_-_91398744
|
14.629
|
|
EPYC
|
epiphycan
|
|
chr3_-_195306274
|
14.605
|
|
APOD
|
apolipoprotein D
|
|
chr1_+_156123321
|
14.577
|
NM_001193300
NM_022367
NM_001193302
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chrX_+_49644469
|
14.567
|
NM_001145073
|
USP27X
|
ubiquitin specific peptidase 27, X-linked
|
|
chr2_+_166152282
|
14.563
|
NM_001040143
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit
|