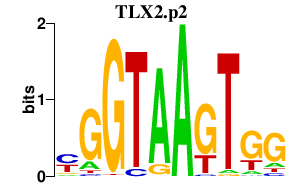
|
chr6_+_111303240
|
22.063
|
NM_032194
|
RPF2
|
ribosome production factor 2 homolog (S. cerevisiae)
|
|
chr12_-_91572328
|
18.513
|
NM_133504
NM_133505
NM_133506
NM_133507
|
DCN
|
decorin
|
|
chr1_-_153363544
|
17.530
|
NM_002964
|
S100A8
|
S100 calcium binding protein A8
|
|
chr19_-_37019169
|
17.038
|
NM_001012756
NM_001166036
NM_001166037
NM_001166038
|
ZNF260
|
zinc finger protein 260
|
|
chr20_+_47895168
|
16.058
|
|
ZNFX1-AS1
|
ZNFX1 antisense RNA 1 (non-protein coding)
|
|
chr16_+_84853585
|
15.223
|
NM_031476
|
CRISPLD2
|
cysteine-rich secretory protein LCCL domain containing 2
|
|
chr15_+_74422920
|
14.654
|
|
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2
|
|
chr20_-_39766522
|
13.431
|
|
|
|
|
chr19_+_58095507
|
12.979
|
NM_001010879
|
ZIK1
|
zinc finger protein interacting with K protein 1 homolog (mouse)
|
|
chr11_+_57365577
|
12.927
|
NM_001032295
|
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1
|
|
chr19_+_37407230
|
12.452
|
NM_001204835
NM_001204836
NM_001204837
NM_001204838
NM_001204839
NM_198539
|
ZNF568
|
zinc finger protein 568
|
|
chr15_+_74422742
|
12.147
|
NM_020851
|
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2
|
|
chr2_+_189839132
|
11.784
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr11_+_117070039
|
11.771
|
NM_001001522
NM_003186
|
TAGLN
|
transgelin
|
|
chr16_+_58283819
|
11.751
|
NM_001142302
NM_014157
|
CCDC113
|
coiled-coil domain containing 113
|
|
chr1_+_153330329
|
11.138
|
NM_002965
|
S100A9
|
S100 calcium binding protein A9
|
|
chr19_+_57752051
|
11.107
|
NM_001023563
NM_001145078
|
ZNF805
|
zinc finger protein 805
|
|
chrX_+_117480416
|
9.887
|
|
WDR44
|
WD repeat domain 44
|
|
chr4_-_88450243
|
9.708
|
|
SPARCL1
|
SPARC-like 1 (hevin)
|
|
chr8_-_27941387
|
9.665
|
NM_001010906
|
C8orf80
|
chromosome 8 open reading frame 80
|
|
chr1_+_196788860
|
9.538
|
NM_002113
|
CFHR1
|
complement factor H-related 1
|
|
chr11_-_124543598
|
9.357
|
NM_170601
|
SIAE
|
sialic acid acetylesterase
|
|
chr2_+_189839113
|
9.319
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr1_-_203320249
|
9.319
|
|
|
|
|
chr12_-_15038724
|
8.967
|
NM_000900
NM_001190839
|
MGP
|
matrix Gla protein
|
|
chr2_-_79315118
|
8.951
|
NM_006507
|
REG1B
|
regenerating islet-derived 1 beta
|
|
chr14_+_23299102
|
8.915
|
|
MRPL52
|
mitochondrial ribosomal protein L52
|
|
chr10_-_44880452
|
8.695
|
NM_000609
NM_001033886
NM_001178134
NM_199168
|
CXCL12
|
chemokine (C-X-C motif) ligand 12
|
|
chr5_+_79783799
|
8.684
|
NM_205548
|
FAM151B
|
family with sequence similarity 151, member B
|
|
chr2_+_189839078
|
8.591
|
NM_000090
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr19_+_859658
|
8.574
|
NM_001928
|
CFD
|
complement factor D (adipsin)
|
|
chr14_+_23299083
|
8.497
|
NM_178336
NM_180982
NM_181304
NM_181305
NM_181306
NM_181307
|
MRPL52
|
mitochondrial ribosomal protein L52
|
|
chr15_-_90347534
|
8.477
|
|
ANPEP
|
alanyl (membrane) aminopeptidase
|
|
chr10_-_90712510
|
8.460
|
NM_001613
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr14_+_101292483
|
8.201
|
|
|
|
|
chr11_+_113930430
|
8.102
|
NM_006006
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr15_+_74422589
|
8.088
|
NM_001130138
|
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2
|
|
chr14_+_101292451
|
7.906
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr1_-_203320185
|
7.883
|
|
FMOD
|
fibromodulin
|
|
chr19_+_41509825
|
7.831
|
|
CYP2B6
|
cytochrome P450, family 2, subfamily B, polypeptide 6
|
|
chr12_-_99038401
|
7.774
|
NM_153687
NM_201612
NM_201613
|
IKBIP
|
IKBKB interacting protein
|
|
chr20_-_43883183
|
7.715
|
NM_003064
|
SLPI
|
secretory leukocyte peptidase inhibitor
|
|
chr3_-_49158171
|
7.603
|
|
USP19
|
ubiquitin specific peptidase 19
|
|
chr17_+_41857802
|
7.586
|
NM_001136483
|
C17orf105
|
chromosome 17 open reading frame 105
|
|
chr14_+_55034633
|
7.418
|
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr16_+_69166538
|
7.382
|
|
CIRH1A
|
cirrhosis, autosomal recessive 1A (cirhin)
|
|
chr3_-_49158256
|
7.325
|
|
USP19
|
ubiquitin specific peptidase 19
|
|
chr7_-_99573637
|
7.259
|
NM_001185
|
AZGP1
|
alpha-2-glycoprotein 1, zinc-binding
|
|
chr12_+_54332575
|
7.199
|
NM_017410
|
HOXC13
|
homeobox C13
|
|
chr3_-_49158357
|
7.131
|
NM_001199160
NM_001199161
NM_001199162
NM_006677
|
USP19
|
ubiquitin specific peptidase 19
|
|
chr9_-_79520878
|
7.123
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr1_-_220445747
|
7.115
|
|
RAB3GAP2
|
RAB3 GTPase activating protein subunit 2 (non-catalytic)
|
|
chr1_-_161279724
|
7.113
|
NM_000530
|
MPZ
|
myelin protein zero
|
|
chr1_+_15764934
|
7.079
|
NM_007272
|
CTRC
|
chymotrypsin C (caldecrin)
|
|
chr4_-_25032306
|
7.074
|
NM_018176
|
LGI2
|
leucine-rich repeat LGI family, member 2
|
|
chr4_-_46911223
|
7.055
|
NM_130902
|
COX7B2
|
cytochrome c oxidase subunit VIIb2
|
|
chr1_-_154193044
|
7.042
|
|
C1orf43
|
chromosome 1 open reading frame 43
|
|
chr2_+_10183630
|
6.947
|
NM_003597
NM_001177716
|
KLF11
|
Kruppel-like factor 11
|
|
chr19_-_6720585
|
6.918
|
NM_000064
|
C3
|
complement component 3
|
|
chr19_+_859673
|
6.771
|
|
CFD
|
complement factor D (adipsin)
|
|
chr6_-_32557431
|
6.757
|
NM_001243965
NM_002124
|
HLA-DRB1
HLA-DRB4
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 1
major histocompatibility complex, class II, DR beta 4
major histocompatibility complex, class II, DR beta 5
|
|
chr12_-_91505215
|
6.700
|
|
LUM
|
lumican
|
|
chr16_+_55512801
|
6.693
|
NM_004530
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr2_-_234475400
|
6.665
|
|
USP40
|
ubiquitin specific peptidase 40
|
|
chr14_+_76618289
|
6.604
|
|
C14orf118
|
chromosome 14 open reading frame 118
|
|
chr3_+_10068123
|
6.520
|
|
FANCD2
|
Fanconi anemia, complementation group D2
|
|
chr1_-_79472360
|
6.410
|
NM_022159
|
ELTD1
|
EGF, latrophilin and seven transmembrane domain containing 1
|
|
chr16_-_72206023
|
6.351
|
NM_031293
|
PMFBP1
|
polyamine modulated factor 1 binding protein 1
|
|
chr8_+_133879202
|
6.309
|
NM_003235
|
TG
|
thyroglobulin
|
|
chr16_+_30386097
|
6.295
|
NM_013292
|
MYLPF
|
myosin light chain, phosphorylatable, fast skeletal muscle
|
|
chr2_+_79347487
|
6.247
|
NM_002909
|
REG1A
|
regenerating islet-derived 1 alpha
|
|
chr8_+_80523376
|
6.188
|
|
STMN2
|
stathmin-like 2
|
|
chr8_+_80523339
|
6.182
|
|
STMN2
|
stathmin-like 2
|
|
chr2_-_160568837
|
6.133
|
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B
|
|
chr8_+_80523304
|
6.098
|
|
STMN2
|
stathmin-like 2
|
|
chr19_+_52264452
|
6.090
|
NM_001005738
|
FPR2
|
formyl peptide receptor 2
|
|
chr8_+_80523351
|
6.062
|
|
STMN2
|
stathmin-like 2
|
|
chr14_-_25078818
|
6.024
|
NM_033423
|
GZMH
|
granzyme H (cathepsin G-like 2, protein h-CCPX)
|
|
chr10_-_99205742
|
5.966
|
NM_016046
|
EXOSC1
|
exosome component 1
|
|
chr6_+_33043702
|
5.908
|
NM_002121
|
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1
|
|
chr3_+_10068097
|
5.866
|
NM_001018115
NM_033084
|
FANCD2
|
Fanconi anemia, complementation group D2
|
|
chr21_+_35736322
|
5.849
|
NM_172201
|
KCNE2
|
potassium voltage-gated channel, Isk-related family, member 2
|
|
chr12_+_32112286
|
5.779
|
NM_018169
|
C12orf35
|
chromosome 12 open reading frame 35
|
|
chr7_-_97881436
|
5.755
|
NM_015395
|
TECPR1
|
tectonin beta-propeller repeat containing 1
|
|
chr1_-_153029987
|
5.733
|
NM_005988
|
SPRR2A
|
small proline-rich protein 2A
|
|
chr3_-_46506342
|
5.712
|
NM_002343
|
LTF
|
lactotransferrin
|
|
chr1_+_1370902
|
5.684
|
NM_022834
NM_199121
|
VWA1
|
von Willebrand factor A domain containing 1
|
|
chr16_+_75252882
|
5.682
|
NM_001906
|
CTRB1
|
chymotrypsinogen B1
|
|
chr18_+_21572736
|
5.666
|
NM_153211
|
TTC39C
|
tetratricopeptide repeat domain 39C
|
|
chr10_-_47173917
|
5.657
|
NM_001040084
NM_001098845
|
ANXA8L2
ANXA8
ANXA8L1
|
annexin A8-like 2
annexin A8
annexin A8-like 1
|
|
chr14_+_76618244
|
5.652
|
NM_017926
NM_017972
|
C14orf118
|
chromosome 14 open reading frame 118
|
|
chr6_-_31125960
|
5.626
|
NM_019052
|
CCHCR1
|
coiled-coil alpha-helical rod protein 1
|
|
chr10_-_75401492
|
5.578
|
NM_021245
|
MYOZ1
|
myozenin 1
|
|
chr9_-_115480363
|
5.544
|
NM_021218
|
C9orf80
|
chromosome 9 open reading frame 80
|
|
chr6_+_32407618
|
5.523
|
NM_019111
|
HLA-DRA
|
major histocompatibility complex, class II, DR alpha
|
|
chr1_-_203055088
|
5.445
|
|
MYOG
|
myogenin (myogenic factor 4)
|
|
chr6_-_99873129
|
5.411
|
NM_015491
NM_032870
|
PNISR
|
PNN-interacting serine/arginine-rich protein
|
|
chr9_-_130477918
|
5.401
|
NM_001002913
|
PTRH1
|
peptidyl-tRNA hydrolase 1 homolog (S. cerevisiae)
|
|
chr5_+_134240820
|
5.386
|
|
PCBD2
|
pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) 2
|
|
chr4_+_74270832
|
5.386
|
|
ALB
|
albumin
|
|
chr19_+_17186569
|
5.375
|
NM_001130065
NM_004145
|
MYO9B
|
myosin IXB
|
|
chr21_-_46340776
|
5.361
|
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit)
|
|
chr12_-_48551269
|
5.352
|
NM_024095
|
ASB8
|
ankyrin repeat and SOCS box containing 8
|
|
chr5_-_140012730
|
5.340
|
|
CD14
|
CD14 molecule
|
|
chr3_+_186330844
|
5.328
|
NM_001622
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr19_-_36236291
|
5.269
|
NM_001040425
NM_144987
|
U2AF1L4
|
U2 small nuclear RNA auxiliary factor 1-like 4
|
|
chr10_-_74114655
|
5.265
|
|
DNAJB12
|
DnaJ (Hsp40) homolog, subfamily B, member 12
|
|
chr1_+_160765927
|
5.263
|
NM_001033667
NM_002348
|
LY9
|
lymphocyte antigen 9
|
|
chr1_-_120935893
|
5.260
|
|
FCGR1B
|
Fc fragment of IgG, high affinity Ib, receptor (CD64)
|
|
chr7_-_43909072
|
5.256
|
NM_032014
|
MRPS24
|
mitochondrial ribosomal protein S24
|
|
chr3_+_186330711
|
5.252
|
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr11_-_64510461
|
5.217
|
|
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated)
|
|
chr3_+_186330859
|
5.207
|
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr1_-_114429996
|
5.189
|
|
BCL2L15
|
BCL2-like 15
|
|
chr14_+_76618272
|
5.164
|
|
C14orf118
|
chromosome 14 open reading frame 118
|
|
chr6_-_32497925
|
5.144
|
NM_002125
|
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 5
|
|
chr21_-_46340821
|
5.144
|
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit)
|
|
chr19_+_33685489
|
5.124
|
NM_002333
|
LRP3
|
low density lipoprotein receptor-related protein 3
|
|
chr1_-_1709863
|
5.116
|
NM_023018
|
NADK
|
NAD kinase
|
|
chr19_+_12721731
|
5.098
|
NM_153358
|
ZNF791
|
zinc finger protein 791
|
|
chr1_-_203320287
|
5.085
|
NM_002023
|
FMOD
|
fibromodulin
|
|
chr17_-_43568081
|
5.064
|
|
PLEKHM1
|
pleckstrin homology domain containing, family M (with RUN domain) member 1
|
|
chr20_+_42839599
|
5.051
|
|
LOC100505783
|
uncharacterized LOC100505783
|
|
chr1_+_145438437
|
5.037
|
NM_006472
|
TXNIP
|
thioredoxin interacting protein
|
|
chr17_-_43138311
|
4.999
|
NM_024819
|
DCAKD
|
dephospho-CoA kinase domain containing
|
|
chr19_+_16999871
|
4.992
|
|
F2RL3
|
coagulation factor II (thrombin) receptor-like 3
|
|
chr6_+_99872769
|
4.966
|
|
|
|
|
chr7_+_80267972
|
4.962
|
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr11_-_64510339
|
4.939
|
|
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated)
|
|
chrX_+_11311532
|
4.938
|
NM_001142
NM_182680
NM_182681
|
AMELX
|
amelogenin, X-linked
|
|
chr20_-_35274557
|
4.924
|
NM_032214
NM_175077
|
SLA2
|
Src-like-adaptor 2
|
|
chr14_-_21271002
|
4.914
|
NM_002933
NM_198234
NM_198235
|
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic)
|
|
chr10_-_115933884
|
4.912
|
|
C10orf118
|
chromosome 10 open reading frame 118
|
|
chr1_-_151762905
|
4.869
|
NM_001083963
NM_001083964
NM_001083965
NM_006862
|
TDRKH
|
tudor and KH domain containing
|
|
chr9_+_132427919
|
4.821
|
NM_016307
|
PRRX2
|
paired related homeobox 2
|
|
chr10_+_49609641
|
4.802
|
NM_002750
NM_139046
NM_139047
NM_139049
|
MAPK8
|
mitogen-activated protein kinase 8
|
|
chr10_-_75401199
|
4.800
|
|
MYOZ1
|
myozenin 1
|
|
chr1_-_161519526
|
4.798
|
|
FCGR3A
|
Fc fragment of IgG, low affinity IIIa, receptor (CD16a)
|
|
chr1_+_33004771
|
4.794
|
NM_001040441
|
ZBTB8A
|
zinc finger and BTB domain containing 8A
|
|
chr2_+_79347610
|
4.755
|
|
REG1A
|
regenerating islet-derived 1 alpha
|
|
chr9_-_88969323
|
4.728
|
NM_001185059
NM_001185074
NM_024617
|
ZCCHC6
|
zinc finger, CCHC domain containing 6
|
|
chr16_+_29674564
|
4.724
|
|
SPN
|
sialophorin
|
|
chr5_-_157002705
|
4.714
|
|
|
|
|
chr10_-_33623299
|
4.709
|
|
NRP1
|
neuropilin 1
|
|
chr1_-_32169555
|
4.709
|
|
COL16A1
|
collagen, type XVI, alpha 1
|
|
chr11_+_2323356
|
4.692
|
|
TSPAN32
|
tetraspanin 32
|
|
chr1_+_36689996
|
4.692
|
NM_005119
|
THRAP3
|
thyroid hormone receptor associated protein 3
|
|
chr5_-_39270708
|
4.675
|
NM_001243093
|
FYB
|
FYN binding protein
|
|
chr2_-_218843541
|
4.660
|
|
TNS1
|
tensin 1
|
|
chr12_+_32654976
|
4.654
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr1_-_31230594
|
4.651
|
|
LAPTM5
|
lysosomal protein transmembrane 5
|
|
chr8_+_17434700
|
4.644
|
|
PDGFRL
|
platelet-derived growth factor receptor-like
|
|
chr6_+_37137882
|
4.641
|
NM_001243186
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr9_-_129870311
|
4.629
|
|
ANGPTL2
|
angiopoietin-like 2
|
|
chr1_+_16174358
|
4.621
|
NM_015001
|
SPEN
|
spen homolog, transcriptional regulator (Drosophila)
|
|
chr18_+_49867069
|
4.607
|
|
DCC
|
deleted in colorectal carcinoma
|
|
chr19_+_11457160
|
4.562
|
NM_001080503
|
CCDC159
|
coiled-coil domain containing 159
|
|
chr7_-_38389424
|
4.561
|
|
|
|
|
chr17_+_47653474
|
4.559
|
|
NXPH3
|
neurexophilin 3
|
|
chr14_-_106110894
|
4.535
|
|
IGHG2
|
immunoglobulin heavy constant gamma 2 (G2m marker)
|
|
chr16_-_72206348
|
4.535
|
NM_001160213
|
PMFBP1
|
polyamine modulated factor 1 binding protein 1
|
|
chr7_-_108166514
|
4.534
|
|
PNPLA8
|
patatin-like phospholipase domain containing 8
|
|
chrX_-_65858882
|
4.477
|
NM_021783
|
EDA2R
|
ectodysplasin A2 receptor
|
|
chr4_-_155533757
|
4.477
|
|
FGG
|
fibrinogen gamma chain
|
|
chr1_-_1590403
|
4.467
|
|
CDK11A
CDK11B
|
cyclin-dependent kinase 11A
cyclin-dependent kinase 11B
|
|
chr14_+_35515347
|
4.467
|
NM_173607
|
FAM177A1
|
family with sequence similarity 177, member A1
|
|
chr6_-_116422408
|
4.464
|
|
|
|
|
chr8_+_22210311
|
4.453
|
|
PIWIL2
|
piwi-like 2 (Drosophila)
|
|
chr17_+_74734106
|
4.448
|
|
MFSD11
|
major facilitator superfamily domain containing 11
|
|
chr3_-_114343791
|
4.444
|
NM_001164346
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr17_-_46667596
|
4.444
|
|
HOXB3
|
homeobox B3
|
|
chr3_+_9404706
|
4.394
|
NM_001114092
NM_015453
|
THUMPD3
|
THUMP domain containing 3
|
|
chr7_-_44105162
|
4.385
|
NM_000290
|
PGAM2
|
phosphoglycerate mutase 2 (muscle)
|
|
chr15_-_77197743
|
4.363
|
|
SCAPER
|
S-phase cyclin A-associated protein in the ER
|
|
chrX_+_67867510
|
4.354
|
NM_001142503
NM_014725
|
STARD8
|
StAR-related lipid transfer (START) domain containing 8
|
|
chrX_+_15518899
|
4.352
|
NM_203281
|
BMX
|
BMX non-receptor tyrosine kinase
|
|
chr4_-_155533800
|
4.341
|
|
FGG
|
fibrinogen gamma chain
|
|
chr9_-_129884885
|
4.333
|
|
ANGPTL2
|
angiopoietin-like 2
|
|
chr9_+_88897112
|
4.331
|
|
|
|
|
chr21_-_46340874
|
4.326
|
NM_000211
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit)
|
|
chr9_+_100395704
|
4.294
|
NM_002486
|
NCBP1
|
nuclear cap binding protein subunit 1, 80kDa
|
|
chr3_-_114343052
|
4.291
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr17_-_43568124
|
4.289
|
NM_014798
|
PLEKHM1
|
pleckstrin homology domain containing, family M (with RUN domain) member 1
|
|
chr2_+_149633173
|
4.282
|
|
|
|
|
chr14_-_53417272
|
4.271
|
|
FERMT2
|
fermitin family member 2
|
|
chr7_+_135347211
|
4.261
|
NM_001130929
|
SLC13A4
C7orf73
|
solute carrier family 13 (sodium/sulfate symporters), member 4
chromosome 7 open reading frame 73
|
|
chr19_-_52133589
|
4.253
|
NM_003830
|
SIGLEC5
|
sialic acid binding Ig-like lectin 5
|
|
chr14_+_70233880
|
4.240
|
|
SRSF5
|
serine/arginine-rich splicing factor 5
|
|
chr6_+_36410800
|
4.239
|
|
KCTD20
|
potassium channel tetramerisation domain containing 20
|
|
chr12_+_120105656
|
4.238
|
NM_006253
|
PRKAB1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit
|
|
chr19_+_34112854
|
4.237
|
NM_001127895
NM_001127896
|
CHST8
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 8
|
|
chr21_+_43639202
|
4.230
|
NM_004915
NM_016818
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1
|
|
chr19_-_38806546
|
4.214
|
NM_001039672
NM_001039673
NM_001145463
|
YIF1B
|
Yip1 interacting factor homolog B (S. cerevisiae)
|
|
chr1_-_157522151
|
4.200
|
NM_001195388
NM_031281
|
FCRL5
|
Fc receptor-like 5
|
|
chr8_+_21915371
|
4.195
|
NM_001114135
|
EPB49
|
erythrocyte membrane protein band 4.9 (dematin)
|
|
chr10_+_48255223
|
4.189
|
NM_001040084
NM_001098845
|
ANXA8L2
ANXA8
ANXA8L1
|
annexin A8-like 2
annexin A8
annexin A8-like 1
|
|
chr4_+_88754120
|
4.165
|
NM_001184695
NM_001184696
NM_001184697
NM_020203
|
MEPE
|
matrix extracellular phosphoglycoprotein
|
|
chr22_-_41032623
|
4.157
|
|
MKL1
|
megakaryoblastic leukemia (translocation) 1
|
|
chr12_-_110271182
|
4.151
|
|
TRPV4
|
transient receptor potential cation channel, subfamily V, member 4
|
|
chr19_+_12305869
|
4.133
|
|
|
|