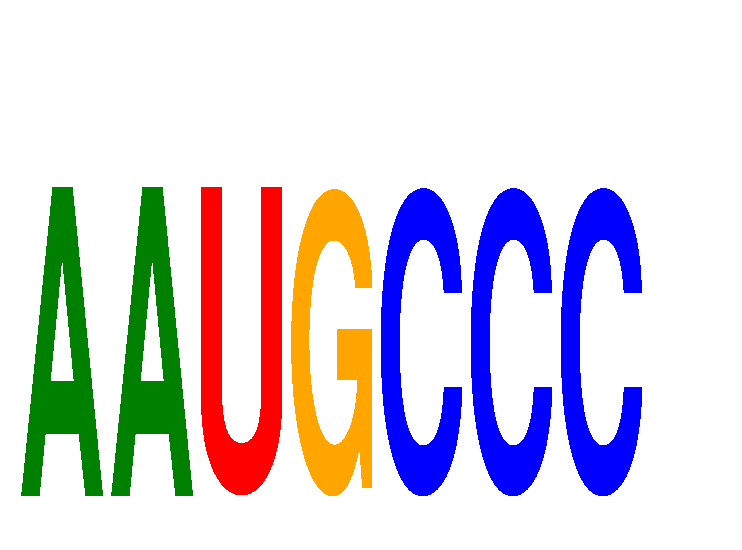Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for AAUGCCC
Z-value: 0.35
miRNA associated with seed AAUGCCC
| Name | miRBASE accession |
|---|---|
|
hsa-miR-365a-3p
|
MIMAT0000710 |
|
hsa-miR-365b-3p
|
MIMAT0022834 |
Activity profile of AAUGCCC motif
Sorted Z-values of AAUGCCC motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_153574480 | 1.62 |
ENST00000410080.1
|
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr3_+_23986748 | 1.61 |
ENST00000312521.4
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr21_-_28217721 | 1.46 |
ENST00000284984.3
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr3_-_55523966 | 1.37 |
ENST00000474267.1
|
WNT5A
|
wingless-type MMTV integration site family, member 5A |
| chr3_-_33481835 | 1.34 |
ENST00000283629.3
|
UBP1
|
upstream binding protein 1 (LBP-1a) |
| chr3_+_113775576 | 1.15 |
ENST00000485050.1
ENST00000281273.4 |
QTRTD1
|
queuine tRNA-ribosyltransferase domain containing 1 |
| chr12_-_15942309 | 1.08 |
ENST00000544064.1
ENST00000543523.1 ENST00000536793.1 |
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr12_+_72666407 | 0.99 |
ENST00000261180.4
|
TRHDE
|
thyrotropin-releasing hormone degrading enzyme |
| chr17_-_78009647 | 0.94 |
ENST00000310924.2
|
TBC1D16
|
TBC1 domain family, member 16 |
| chr12_+_66217911 | 0.91 |
ENST00000403681.2
|
HMGA2
|
high mobility group AT-hook 2 |
| chr19_+_46850251 | 0.88 |
ENST00000012443.4
|
PPP5C
|
protein phosphatase 5, catalytic subunit |
| chr5_+_112312416 | 0.87 |
ENST00000389063.2
|
DCP2
|
decapping mRNA 2 |
| chr12_+_68042495 | 0.82 |
ENST00000344096.3
|
DYRK2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr1_+_2160134 | 0.81 |
ENST00000378536.4
|
SKI
|
v-ski avian sarcoma viral oncogene homolog |
| chr4_-_74124502 | 0.81 |
ENST00000358602.4
ENST00000330838.6 ENST00000561029.1 |
ANKRD17
|
ankyrin repeat domain 17 |
| chr16_+_67063036 | 0.80 |
ENST00000290858.6
ENST00000564034.1 |
CBFB
|
core-binding factor, beta subunit |
| chr9_-_14314066 | 0.77 |
ENST00000397575.3
|
NFIB
|
nuclear factor I/B |
| chr3_+_171758344 | 0.73 |
ENST00000336824.4
ENST00000423424.1 |
FNDC3B
|
fibronectin type III domain containing 3B |
| chr6_-_134639180 | 0.72 |
ENST00000367858.5
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr1_-_22109682 | 0.71 |
ENST00000400301.1
ENST00000532737.1 |
USP48
|
ubiquitin specific peptidase 48 |
| chr9_-_117880477 | 0.71 |
ENST00000534839.1
ENST00000340094.3 ENST00000535648.1 ENST00000346706.3 ENST00000345230.3 ENST00000350763.4 |
TNC
|
tenascin C |
| chr9_+_114659046 | 0.66 |
ENST00000374279.3
|
UGCG
|
UDP-glucose ceramide glucosyltransferase |
| chr3_-_79068594 | 0.66 |
ENST00000436010.2
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr12_-_31479045 | 0.65 |
ENST00000539409.1
ENST00000395766.1 |
FAM60A
|
family with sequence similarity 60, member A |
| chr3_-_113465065 | 0.64 |
ENST00000497255.1
ENST00000478020.1 ENST00000240922.3 ENST00000493900.1 |
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr3_-_123603137 | 0.63 |
ENST00000360304.3
ENST00000359169.1 ENST00000346322.5 ENST00000360772.3 |
MYLK
|
myosin light chain kinase |
| chr11_-_31839488 | 0.63 |
ENST00000419022.1
ENST00000379132.3 ENST00000379129.2 |
PAX6
|
paired box 6 |
| chrX_-_109561294 | 0.61 |
ENST00000372059.2
ENST00000262844.5 |
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr2_-_64881018 | 0.61 |
ENST00000313349.3
|
SERTAD2
|
SERTA domain containing 2 |
| chr1_-_153588334 | 0.59 |
ENST00000476873.1
|
S100A14
|
S100 calcium binding protein A14 |
| chrX_+_77359671 | 0.59 |
ENST00000373316.4
|
PGK1
|
phosphoglycerate kinase 1 |
| chr1_-_155881156 | 0.55 |
ENST00000539040.1
ENST00000368323.3 |
RIT1
|
Ras-like without CAAX 1 |
| chr6_-_111804393 | 0.55 |
ENST00000368802.3
ENST00000368805.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr3_-_47823298 | 0.51 |
ENST00000254480.5
|
SMARCC1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 |
| chr5_-_43313574 | 0.51 |
ENST00000325110.6
ENST00000433297.2 |
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr2_-_208634287 | 0.50 |
ENST00000295417.3
|
FZD5
|
frizzled family receptor 5 |
| chr16_+_53088885 | 0.50 |
ENST00000566029.1
ENST00000447540.1 |
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr5_+_133861790 | 0.49 |
ENST00000395003.1
|
PHF15
|
jade family PHD finger 2 |
| chr2_-_183903133 | 0.48 |
ENST00000361354.4
|
NCKAP1
|
NCK-associated protein 1 |
| chr12_-_75905374 | 0.48 |
ENST00000438169.2
ENST00000229214.4 |
KRR1
|
KRR1, small subunit (SSU) processome component, homolog (yeast) |
| chr2_+_46926048 | 0.47 |
ENST00000306503.5
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr10_-_98346801 | 0.47 |
ENST00000371142.4
|
TM9SF3
|
transmembrane 9 superfamily member 3 |
| chr2_-_200322723 | 0.46 |
ENST00000417098.1
|
SATB2
|
SATB homeobox 2 |
| chr5_+_74632993 | 0.46 |
ENST00000287936.4
|
HMGCR
|
3-hydroxy-3-methylglutaryl-CoA reductase |
| chr1_-_205719295 | 0.43 |
ENST00000367142.4
|
NUCKS1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr15_+_96873921 | 0.41 |
ENST00000394166.3
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr1_-_244013384 | 0.40 |
ENST00000366539.1
|
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 |
| chr9_+_131445928 | 0.39 |
ENST00000372692.4
|
SET
|
SET nuclear oncogene |
| chr7_-_27205136 | 0.38 |
ENST00000396345.1
ENST00000343483.6 |
HOXA9
|
homeobox A9 |
| chr11_+_2466218 | 0.38 |
ENST00000155840.5
|
KCNQ1
|
potassium voltage-gated channel, KQT-like subfamily, member 1 |
| chr20_+_56884752 | 0.37 |
ENST00000244040.3
|
RAB22A
|
RAB22A, member RAS oncogene family |
| chr8_+_54764346 | 0.36 |
ENST00000297313.3
ENST00000344277.6 |
RGS20
|
regulator of G-protein signaling 20 |
| chr7_-_32931387 | 0.36 |
ENST00000304056.4
|
KBTBD2
|
kelch repeat and BTB (POZ) domain containing 2 |
| chr14_+_55518349 | 0.35 |
ENST00000395468.4
|
MAPK1IP1L
|
mitogen-activated protein kinase 1 interacting protein 1-like |
| chr8_-_57123815 | 0.35 |
ENST00000316981.3
ENST00000423799.2 ENST00000429357.2 |
PLAG1
|
pleiomorphic adenoma gene 1 |
| chr1_+_36396677 | 0.33 |
ENST00000373191.4
ENST00000397828.2 |
AGO3
|
argonaute RISC catalytic component 3 |
| chr1_-_38325256 | 0.33 |
ENST00000373036.4
|
MTF1
|
metal-regulatory transcription factor 1 |
| chr11_-_27528301 | 0.33 |
ENST00000524596.1
ENST00000278193.2 |
LIN7C
|
lin-7 homolog C (C. elegans) |
| chr12_+_56137064 | 0.31 |
ENST00000257868.5
ENST00000546799.1 |
GDF11
|
growth differentiation factor 11 |
| chr7_+_138145076 | 0.30 |
ENST00000343526.4
|
TRIM24
|
tripartite motif containing 24 |
| chrX_+_23352133 | 0.29 |
ENST00000379361.4
|
PTCHD1
|
patched domain containing 1 |
| chr11_+_64073699 | 0.29 |
ENST00000405666.1
ENST00000468670.1 |
ESRRA
|
estrogen-related receptor alpha |
| chr10_+_22610124 | 0.29 |
ENST00000376663.3
|
BMI1
|
BMI1 polycomb ring finger oncogene |
| chr3_-_15901278 | 0.28 |
ENST00000399451.2
|
ANKRD28
|
ankyrin repeat domain 28 |
| chr18_-_60987220 | 0.28 |
ENST00000398117.1
|
BCL2
|
B-cell CLL/lymphoma 2 |
| chr3_-_125094093 | 0.28 |
ENST00000484491.1
ENST00000492394.1 ENST00000471196.1 ENST00000468369.1 ENST00000544464.1 ENST00000485866.1 ENST00000360647.4 |
ZNF148
|
zinc finger protein 148 |
| chr7_-_83824169 | 0.27 |
ENST00000265362.4
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr3_-_11762202 | 0.26 |
ENST00000445411.1
ENST00000404339.1 ENST00000273038.3 |
VGLL4
|
vestigial like 4 (Drosophila) |
| chr19_-_49576198 | 0.25 |
ENST00000221444.1
|
KCNA7
|
potassium voltage-gated channel, shaker-related subfamily, member 7 |
| chr11_+_34642656 | 0.25 |
ENST00000257831.3
ENST00000450654.2 |
EHF
|
ets homologous factor |
| chr17_-_16118835 | 0.24 |
ENST00000582357.1
ENST00000436828.1 ENST00000411510.1 ENST00000268712.3 |
NCOR1
|
nuclear receptor corepressor 1 |
| chr1_+_29063271 | 0.24 |
ENST00000373812.3
|
YTHDF2
|
YTH domain family, member 2 |
| chr10_-_105615164 | 0.23 |
ENST00000355946.2
ENST00000369774.4 |
SH3PXD2A
|
SH3 and PX domains 2A |
| chr15_+_52121822 | 0.23 |
ENST00000558455.1
ENST00000308580.7 |
TMOD3
|
tropomodulin 3 (ubiquitous) |
| chr4_+_38665810 | 0.23 |
ENST00000261438.5
ENST00000514033.1 |
KLF3
|
Kruppel-like factor 3 (basic) |
| chr3_+_51575596 | 0.22 |
ENST00000409535.2
|
RAD54L2
|
RAD54-like 2 (S. cerevisiae) |
| chr3_-_48885228 | 0.22 |
ENST00000454963.1
ENST00000296446.8 ENST00000419216.1 ENST00000265563.8 |
PRKAR2A
|
protein kinase, cAMP-dependent, regulatory, type II, alpha |
| chr11_-_77532050 | 0.22 |
ENST00000308488.6
|
RSF1
|
remodeling and spacing factor 1 |
| chr11_-_57283159 | 0.21 |
ENST00000533263.1
ENST00000278426.3 |
SLC43A1
|
solute carrier family 43 (amino acid system L transporter), member 1 |
| chr16_+_70557685 | 0.19 |
ENST00000302516.5
ENST00000566095.2 ENST00000577085.1 ENST00000567654.1 |
SF3B3
|
splicing factor 3b, subunit 3, 130kDa |
| chr2_-_157189180 | 0.19 |
ENST00000539077.1
ENST00000424077.1 ENST00000426264.1 ENST00000339562.4 ENST00000421709.1 |
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr2_+_131862900 | 0.19 |
ENST00000438882.2
ENST00000538982.1 ENST00000404460.1 |
PLEKHB2
|
pleckstrin homology domain containing, family B (evectins) member 2 |
| chr21_+_38071430 | 0.18 |
ENST00000290399.6
|
SIM2
|
single-minded family bHLH transcription factor 2 |
| chr18_+_56338618 | 0.17 |
ENST00000348428.3
|
MALT1
|
mucosa associated lymphoid tissue lymphoma translocation gene 1 |
| chr17_+_28804380 | 0.16 |
ENST00000225724.5
ENST00000451249.2 ENST00000467337.2 ENST00000581721.1 ENST00000414833.2 |
GOSR1
|
golgi SNAP receptor complex member 1 |
| chr17_-_56084578 | 0.16 |
ENST00000582730.2
ENST00000584773.1 ENST00000585096.1 ENST00000258962.4 |
SRSF1
|
serine/arginine-rich splicing factor 1 |
| chr13_-_21476900 | 0.15 |
ENST00000400602.2
ENST00000255305.6 |
XPO4
|
exportin 4 |
| chr1_-_78225482 | 0.15 |
ENST00000524778.1
ENST00000370794.3 ENST00000370793.1 ENST00000370792.3 |
USP33
|
ubiquitin specific peptidase 33 |
| chr2_+_192542850 | 0.15 |
ENST00000410026.2
|
NABP1
|
nucleic acid binding protein 1 |
| chr7_+_139044621 | 0.15 |
ENST00000354926.4
|
C7orf55-LUC7L2
|
C7orf55-LUC7L2 readthrough |
| chrX_-_3631635 | 0.14 |
ENST00000262848.5
|
PRKX
|
protein kinase, X-linked |
| chr1_-_155211017 | 0.14 |
ENST00000536770.1
ENST00000368373.3 |
GBA
|
glucosidase, beta, acid |
| chr7_-_151217001 | 0.14 |
ENST00000262187.5
|
RHEB
|
Ras homolog enriched in brain |
| chr7_+_94285637 | 0.13 |
ENST00000482108.1
ENST00000488574.1 |
PEG10
|
paternally expressed 10 |
| chrX_-_74376108 | 0.13 |
ENST00000339447.4
ENST00000373394.3 ENST00000529949.1 ENST00000534524.1 ENST00000253577.3 |
ABCB7
|
ATP-binding cassette, sub-family B (MDR/TAP), member 7 |
| chr4_+_139936905 | 0.12 |
ENST00000280614.2
|
CCRN4L
|
CCR4 carbon catabolite repression 4-like (S. cerevisiae) |
| chr4_-_149365827 | 0.12 |
ENST00000344721.4
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr3_+_101292939 | 0.12 |
ENST00000265260.3
ENST00000469941.1 ENST00000296024.5 |
PCNP
|
PEST proteolytic signal containing nuclear protein |
| chrX_+_69664706 | 0.12 |
ENST00000194900.4
ENST00000374360.3 |
DLG3
|
discs, large homolog 3 (Drosophila) |
| chr13_+_114238997 | 0.11 |
ENST00000538138.1
ENST00000375370.5 |
TFDP1
|
transcription factor Dp-1 |
| chr1_+_161284047 | 0.11 |
ENST00000367975.2
ENST00000342751.4 ENST00000432287.2 ENST00000392169.2 ENST00000513009.1 |
SDHC
|
succinate dehydrogenase complex, subunit C, integral membrane protein, 15kDa |
| chr5_-_142783175 | 0.11 |
ENST00000231509.3
ENST00000394464.2 |
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr15_+_41523335 | 0.11 |
ENST00000334660.5
|
CHP1
|
calcineurin-like EF-hand protein 1 |
| chr2_-_240322643 | 0.10 |
ENST00000345617.3
|
HDAC4
|
histone deacetylase 4 |
| chr1_+_64239657 | 0.10 |
ENST00000371080.1
ENST00000371079.1 |
ROR1
|
receptor tyrosine kinase-like orphan receptor 1 |
| chr16_-_79634595 | 0.10 |
ENST00000326043.4
ENST00000393350.1 |
MAF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog |
| chr1_+_78470530 | 0.10 |
ENST00000370763.5
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4 |
| chrX_+_73641286 | 0.10 |
ENST00000587091.1
|
SLC16A2
|
solute carrier family 16, member 2 (thyroid hormone transporter) |
| chr14_-_21905395 | 0.09 |
ENST00000430710.3
ENST00000553283.1 |
CHD8
|
chromodomain helicase DNA binding protein 8 |
| chrX_-_129244655 | 0.09 |
ENST00000335997.7
|
ELF4
|
E74-like factor 4 (ets domain transcription factor) |
| chr1_-_35658736 | 0.09 |
ENST00000357214.5
|
SFPQ
|
splicing factor proline/glutamine-rich |
| chr8_-_38239732 | 0.08 |
ENST00000534155.1
ENST00000433384.2 ENST00000317025.8 ENST00000316985.3 |
WHSC1L1
|
Wolf-Hirschhorn syndrome candidate 1-like 1 |
| chr8_+_28351707 | 0.08 |
ENST00000537916.1
ENST00000523546.1 ENST00000240093.3 |
FZD3
|
frizzled family receptor 3 |
| chr1_-_39339777 | 0.08 |
ENST00000397572.2
|
MYCBP
|
MYC binding protein |
| chr14_-_99737565 | 0.08 |
ENST00000357195.3
|
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein) |
| chr1_+_203274639 | 0.07 |
ENST00000290551.4
|
BTG2
|
BTG family, member 2 |
| chr9_-_127905736 | 0.07 |
ENST00000336505.6
ENST00000373549.4 |
SCAI
|
suppressor of cancer cell invasion |
| chr1_+_145611010 | 0.07 |
ENST00000369291.5
|
RNF115
|
ring finger protein 115 |
| chr2_+_155554797 | 0.06 |
ENST00000295101.2
|
KCNJ3
|
potassium inwardly-rectifying channel, subfamily J, member 3 |
| chr5_-_168006591 | 0.06 |
ENST00000239231.6
|
PANK3
|
pantothenate kinase 3 |
| chr10_+_101419187 | 0.06 |
ENST00000370489.4
|
ENTPD7
|
ectonucleoside triphosphate diphosphohydrolase 7 |
| chr12_-_67072714 | 0.06 |
ENST00000545666.1
ENST00000398016.3 ENST00000359742.4 ENST00000286445.7 ENST00000538211.1 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr9_+_128024067 | 0.06 |
ENST00000461379.1
ENST00000394084.1 ENST00000394105.2 ENST00000470056.1 ENST00000394104.2 ENST00000265956.4 ENST00000394083.2 ENST00000495955.1 ENST00000467750.1 ENST00000297933.6 |
GAPVD1
|
GTPase activating protein and VPS9 domains 1 |
| chr8_+_21946681 | 0.05 |
ENST00000289921.7
|
FAM160B2
|
family with sequence similarity 160, member B2 |
| chr10_-_120101804 | 0.05 |
ENST00000369183.4
ENST00000369172.4 |
FAM204A
|
family with sequence similarity 204, member A |
| chr4_+_41992489 | 0.05 |
ENST00000264451.7
|
SLC30A9
|
solute carrier family 30 (zinc transporter), member 9 |
| chr22_+_37309662 | 0.05 |
ENST00000403662.3
ENST00000262825.5 |
CSF2RB
|
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) |
| chr3_-_48632593 | 0.05 |
ENST00000454817.1
ENST00000328333.8 |
COL7A1
|
collagen, type VII, alpha 1 |
| chr12_-_89918522 | 0.05 |
ENST00000529983.2
|
GALNT4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) |
| chr6_+_34857019 | 0.05 |
ENST00000360359.3
ENST00000535627.1 |
ANKS1A
|
ankyrin repeat and sterile alpha motif domain containing 1A |
| chr3_+_38495333 | 0.04 |
ENST00000352511.4
|
ACVR2B
|
activin A receptor, type IIB |
| chr3_+_43328004 | 0.03 |
ENST00000454177.1
ENST00000429705.2 ENST00000296088.7 ENST00000437827.1 |
SNRK
|
SNF related kinase |
| chr16_-_67840442 | 0.03 |
ENST00000536251.1
ENST00000448631.2 ENST00000602677.1 ENST00000411657.2 ENST00000425512.2 ENST00000317506.3 |
RANBP10
|
RAN binding protein 10 |
| chr7_+_4721885 | 0.02 |
ENST00000328914.4
|
FOXK1
|
forkhead box K1 |
| chr20_+_60697480 | 0.02 |
ENST00000370915.1
ENST00000253001.4 ENST00000400318.2 ENST00000279068.6 ENST00000279069.7 |
LSM14B
|
LSM14B, SCD6 homolog B (S. cerevisiae) |
| chr2_+_134877740 | 0.02 |
ENST00000409645.1
|
MGAT5
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
| chr18_+_43753974 | 0.01 |
ENST00000282059.6
ENST00000321319.6 |
C18orf25
|
chromosome 18 open reading frame 25 |
Network of associatons between targets according to the STRING database.
First level regulatory network of AAUGCCC
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:1904933 | cardiac right atrium morphogenesis(GO:0003213) negative regulation of melanin biosynthetic process(GO:0048022) positive regulation of anagen(GO:0051885) mediolateral intercalation(GO:0060031) cervix development(GO:0060067) lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) planar cell polarity pathway involved in gastrula mediolateral intercalation(GO:0060775) negative regulation of secondary metabolite biosynthetic process(GO:1900377) regulation of cell proliferation in midbrain(GO:1904933) |
| 0.3 | 0.9 | GO:2000685 | mesodermal-endodermal cell signaling(GO:0003131) programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) histone H2A-S139 phosphorylation(GO:0035978) positive regulation of cellular response to X-ray(GO:2000685) |
| 0.3 | 1.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.2 | 0.7 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.2 | 0.7 | GO:0006679 | glucosylceramide biosynthetic process(GO:0006679) |
| 0.2 | 0.7 | GO:0050923 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 0.2 | 0.8 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.2 | 0.8 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.2 | 0.5 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.2 | 0.8 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 0.9 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 0.6 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.1 | 0.9 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.8 | GO:2000795 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 0.4 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.1 | 0.7 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.4 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.1 | 1.5 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.1 | 0.6 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.1 | 0.5 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 0.6 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.1 | GO:0045976 | negative regulation of mitotic cell cycle, embryonic(GO:0045976) |
| 0.1 | 0.3 | GO:0043375 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.1 | 0.3 | GO:0048880 | sensory system development(GO:0048880) |
| 0.1 | 0.2 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.1 | 0.2 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.1 | 1.6 | GO:2000505 | regulation of energy homeostasis(GO:2000505) regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.1 | 0.2 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.1 | 0.5 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.5 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.2 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing by siRNA(GO:0090625) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.4 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.0 | 0.5 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.3 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.5 | GO:0071372 | response to light intensity(GO:0009642) cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:1901804 | beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.9 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.2 | GO:1903677 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 | 0.4 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.3 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.0 | 0.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.6 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 0.4 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.8 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.0 | 1.0 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.6 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.9 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 0.1 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.3 | GO:0021511 | spinal cord patterning(GO:0021511) |
| 0.0 | 0.2 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.0 | 0.2 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.3 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 1.6 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.0 | 0.1 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.1 | GO:0070885 | positive regulation of sodium:proton antiporter activity(GO:0032417) negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 | 0.1 | GO:0099624 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) atrial cardiac muscle cell membrane repolarization(GO:0099624) |
| 0.0 | 0.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.1 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) peptidyl-lysine deacetylation(GO:0034983) cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.0 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.1 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.6 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.9 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 0.2 | GO:0031213 | RSF complex(GO:0031213) |
| 0.1 | 1.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 1.6 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 1.1 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 0.8 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 0.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.3 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.2 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.6 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.5 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.8 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.2 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.2 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 1.9 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.1 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.5 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 1.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.3 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0031588 | cAMP-dependent protein kinase complex(GO:0005952) nucleotide-activated protein kinase complex(GO:0031588) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0035501 | MH1 domain binding(GO:0035501) |
| 0.3 | 0.8 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.2 | 0.6 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.1 | 1.6 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 0.6 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.7 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.5 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 0.9 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.1 | 0.4 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.1 | 0.7 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 1.4 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 0.3 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.1 | 0.7 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.6 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 1.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 1.0 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.0 | 0.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.7 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.3 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.1 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.1 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.2 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.9 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.5 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.0 | 1.2 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.8 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.2 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 1.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.2 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.7 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.6 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.1 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.7 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.6 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 2.2 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.6 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.4 | ST G ALPHA I PATHWAY | G alpha i Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 1.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 2.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.8 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.6 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 1.0 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.2 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.0 | 0.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.4 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |