Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
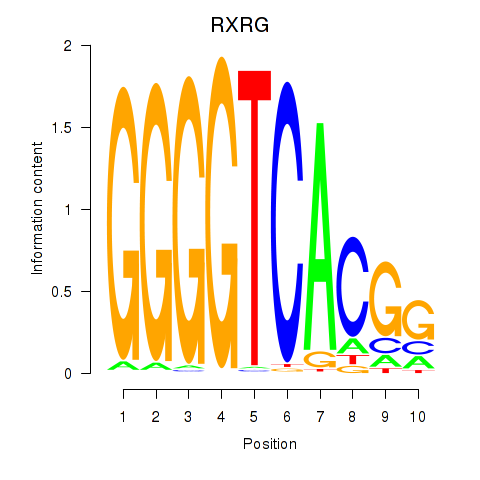
Results for RXRG
Z-value: 1.12
Transcription factors associated with RXRG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RXRG
|
ENSG00000143171.8 | retinoid X receptor gamma |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RXRG | hg19_v2_chr1_-_165414414_165414433 | 0.18 | 8.9e-03 | Click! |
Activity profile of RXRG motif
Sorted Z-values of RXRG motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_27278304 | 17.86 |
ENST00000577226.1
|
PHF12
|
PHD finger protein 12 |
| chr16_+_222846 | 12.78 |
ENST00000251595.6
ENST00000397806.1 |
HBA2
|
hemoglobin, alpha 2 |
| chr12_-_54982420 | 9.37 |
ENST00000257905.8
|
PPP1R1A
|
protein phosphatase 1, regulatory (inhibitor) subunit 1A |
| chr8_-_70983506 | 9.15 |
ENST00000276594.2
|
PRDM14
|
PR domain containing 14 |
| chr17_+_7211280 | 9.11 |
ENST00000419711.2
ENST00000571955.1 ENST00000573714.1 |
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr9_+_139873264 | 8.52 |
ENST00000446677.1
|
PTGDS
|
prostaglandin D2 synthase 21kDa (brain) |
| chr12_-_54982300 | 8.40 |
ENST00000547431.1
|
PPP1R1A
|
protein phosphatase 1, regulatory (inhibitor) subunit 1A |
| chr19_-_1174226 | 8.38 |
ENST00000587024.1
ENST00000361757.3 |
SBNO2
|
strawberry notch homolog 2 (Drosophila) |
| chr11_+_125774362 | 7.45 |
ENST00000530414.1
ENST00000530129.2 |
DDX25
|
DEAD (Asp-Glu-Ala-Asp) box helicase 25 |
| chr20_-_44539538 | 7.43 |
ENST00000372420.1
|
PLTP
|
phospholipid transfer protein |
| chr19_+_35630022 | 7.27 |
ENST00000589209.1
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr19_-_14228541 | 7.23 |
ENST00000590853.1
ENST00000308677.4 |
PRKACA
|
protein kinase, cAMP-dependent, catalytic, alpha |
| chr19_-_39390440 | 7.05 |
ENST00000249396.7
ENST00000414941.1 ENST00000392081.2 |
SIRT2
|
sirtuin 2 |
| chrX_+_38420783 | 6.90 |
ENST00000422612.2
ENST00000286824.6 ENST00000545599.1 |
TSPAN7
|
tetraspanin 7 |
| chr6_+_107811162 | 6.90 |
ENST00000317357.5
|
SOBP
|
sine oculis binding protein homolog (Drosophila) |
| chr19_-_39390350 | 6.56 |
ENST00000447739.1
ENST00000358931.5 ENST00000407552.1 |
SIRT2
|
sirtuin 2 |
| chr22_+_18593446 | 6.54 |
ENST00000316027.6
|
TUBA8
|
tubulin, alpha 8 |
| chr10_+_76586348 | 6.49 |
ENST00000372724.1
ENST00000287239.4 ENST00000372714.1 |
KAT6B
|
K(lysine) acetyltransferase 6B |
| chr22_+_19701985 | 6.16 |
ENST00000455784.2
ENST00000406395.1 |
SEPT5
|
septin 5 |
| chr17_+_42264322 | 5.92 |
ENST00000446571.3
ENST00000357984.3 ENST00000538716.2 |
TMUB2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr7_-_35734730 | 5.80 |
ENST00000396081.1
ENST00000311350.3 |
HERPUD2
|
HERPUD family member 2 |
| chr3_+_133465228 | 5.80 |
ENST00000482271.1
ENST00000264998.3 |
TF
|
transferrin |
| chr17_+_42264395 | 5.77 |
ENST00000587989.1
ENST00000590235.1 |
TMUB2
|
transmembrane and ubiquitin-like domain containing 2 |
| chrX_+_38420623 | 5.68 |
ENST00000378482.2
|
TSPAN7
|
tetraspanin 7 |
| chr20_-_35492048 | 5.44 |
ENST00000237536.4
|
SOGA1
|
suppressor of glucose, autophagy associated 1 |
| chr6_-_52859968 | 5.41 |
ENST00000370959.1
|
GSTA4
|
glutathione S-transferase alpha 4 |
| chrX_+_44732757 | 5.35 |
ENST00000377967.4
ENST00000536777.1 ENST00000382899.4 ENST00000543216.1 |
KDM6A
|
lysine (K)-specific demethylase 6A |
| chr5_-_176056974 | 5.27 |
ENST00000510387.1
ENST00000506696.1 |
SNCB
|
synuclein, beta |
| chr2_+_105471969 | 5.18 |
ENST00000361360.2
|
POU3F3
|
POU class 3 homeobox 3 |
| chr3_+_49591881 | 5.18 |
ENST00000296452.4
|
BSN
|
bassoon presynaptic cytomatrix protein |
| chrX_+_55478538 | 5.18 |
ENST00000342972.1
|
MAGEH1
|
melanoma antigen family H, 1 |
| chr19_+_35630628 | 5.17 |
ENST00000588715.1
ENST00000588607.1 |
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr10_-_81205373 | 5.17 |
ENST00000372336.3
|
ZCCHC24
|
zinc finger, CCHC domain containing 24 |
| chr1_+_202317815 | 5.15 |
ENST00000608999.1
ENST00000336894.4 ENST00000480184.1 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr22_-_38699003 | 5.14 |
ENST00000451964.1
|
CSNK1E
|
casein kinase 1, epsilon |
| chr19_-_46000251 | 5.09 |
ENST00000590526.1
ENST00000344680.4 ENST00000245923.4 |
RTN2
|
reticulon 2 |
| chr19_+_58095501 | 4.97 |
ENST00000536878.2
ENST00000597850.1 ENST00000597219.1 ENST00000598689.1 ENST00000599456.1 ENST00000307468.4 |
ZIK1
|
zinc finger protein interacting with K protein 1 |
| chr7_+_100136811 | 4.96 |
ENST00000300176.4
ENST00000262935.4 |
AGFG2
|
ArfGAP with FG repeats 2 |
| chr8_+_144816303 | 4.96 |
ENST00000533004.1
|
FAM83H-AS1
|
FAM83H antisense RNA 1 (head to head) |
| chr1_-_32210275 | 4.91 |
ENST00000440175.2
|
BAI2
|
brain-specific angiogenesis inhibitor 2 |
| chr7_-_45960850 | 4.85 |
ENST00000381083.4
ENST00000381086.5 ENST00000275521.6 |
IGFBP3
|
insulin-like growth factor binding protein 3 |
| chr7_+_30068260 | 4.78 |
ENST00000440706.2
|
PLEKHA8
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 8 |
| chr1_+_44412577 | 4.68 |
ENST00000372343.3
|
IPO13
|
importin 13 |
| chr1_-_38512450 | 4.66 |
ENST00000373012.2
|
POU3F1
|
POU class 3 homeobox 1 |
| chr22_+_19705928 | 4.62 |
ENST00000383045.3
ENST00000438754.2 |
SEPT5
|
septin 5 |
| chr19_+_52693259 | 4.62 |
ENST00000322088.6
ENST00000454220.2 ENST00000444322.2 ENST00000477989.1 |
PPP2R1A
|
protein phosphatase 2, regulatory subunit A, alpha |
| chr19_+_50354430 | 4.60 |
ENST00000599732.1
|
PTOV1
|
prostate tumor overexpressed 1 |
| chr17_+_3627185 | 4.59 |
ENST00000325418.4
|
GSG2
|
germ cell associated 2 (haspin) |
| chr19_-_42746714 | 4.57 |
ENST00000222330.3
|
GSK3A
|
glycogen synthase kinase 3 alpha |
| chr8_-_57359131 | 4.56 |
ENST00000518974.1
ENST00000523051.1 ENST00000518770.1 ENST00000451791.2 |
PENK
|
proenkephalin |
| chr19_-_1605424 | 4.56 |
ENST00000589880.1
ENST00000585671.1 ENST00000591899.3 |
UQCR11
|
ubiquinol-cytochrome c reductase, complex III subunit XI |
| chr9_-_123342415 | 4.53 |
ENST00000349780.4
ENST00000360190.4 ENST00000360822.3 ENST00000359309.3 |
CDK5RAP2
|
CDK5 regulatory subunit associated protein 2 |
| chr11_-_86666427 | 4.49 |
ENST00000531380.1
|
FZD4
|
frizzled family receptor 4 |
| chr19_+_50354393 | 4.48 |
ENST00000391842.1
|
PTOV1
|
prostate tumor overexpressed 1 |
| chr5_-_132112921 | 4.45 |
ENST00000378721.4
ENST00000378701.1 |
SEPT8
|
septin 8 |
| chr6_-_46459099 | 4.37 |
ENST00000371374.1
|
RCAN2
|
regulator of calcineurin 2 |
| chr19_+_50354462 | 4.36 |
ENST00000601675.1
|
PTOV1
|
prostate tumor overexpressed 1 |
| chr5_-_132112907 | 4.35 |
ENST00000458488.2
|
SEPT8
|
septin 8 |
| chr2_+_46524537 | 4.27 |
ENST00000263734.3
|
EPAS1
|
endothelial PAS domain protein 1 |
| chr5_+_149546334 | 4.26 |
ENST00000231656.8
|
CDX1
|
caudal type homeobox 1 |
| chr1_-_156399184 | 4.24 |
ENST00000368243.1
ENST00000357975.4 ENST00000310027.5 ENST00000400991.2 |
C1orf61
|
chromosome 1 open reading frame 61 |
| chr3_-_28390581 | 4.21 |
ENST00000479665.1
|
AZI2
|
5-azacytidine induced 2 |
| chr3_-_50340996 | 4.21 |
ENST00000266031.4
ENST00000395143.2 ENST00000457214.2 ENST00000447605.2 ENST00000418723.1 ENST00000395144.2 |
HYAL1
|
hyaluronoglucosaminidase 1 |
| chr17_-_40575535 | 4.21 |
ENST00000357037.5
|
PTRF
|
polymerase I and transcript release factor |
| chr19_+_54606145 | 4.18 |
ENST00000485876.1
ENST00000391762.1 ENST00000471292.1 ENST00000391763.3 ENST00000391764.3 ENST00000303553.5 |
NDUFA3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3, 9kDa |
| chrX_+_135229600 | 4.18 |
ENST00000370690.3
|
FHL1
|
four and a half LIM domains 1 |
| chr22_+_20104947 | 4.18 |
ENST00000402752.1
|
RANBP1
|
RAN binding protein 1 |
| chr22_-_20104700 | 4.17 |
ENST00000439169.2
ENST00000445045.1 ENST00000404751.3 ENST00000252136.7 ENST00000403707.3 |
TRMT2A
|
tRNA methyltransferase 2 homolog A (S. cerevisiae) |
| chr17_-_38256973 | 4.17 |
ENST00000246672.3
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1 |
| chr19_-_39322497 | 4.14 |
ENST00000221418.4
|
ECH1
|
enoyl CoA hydratase 1, peroxisomal |
| chr17_-_79139817 | 4.11 |
ENST00000326724.4
|
AATK
|
apoptosis-associated tyrosine kinase |
| chr19_+_7968728 | 4.10 |
ENST00000397981.3
ENST00000545011.1 ENST00000397983.3 ENST00000397979.3 |
MAP2K7
|
mitogen-activated protein kinase kinase 7 |
| chrX_+_129473859 | 4.10 |
ENST00000424447.1
|
SLC25A14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr16_-_70719925 | 4.08 |
ENST00000338779.6
|
MTSS1L
|
metastasis suppressor 1-like |
| chr9_+_137533615 | 4.06 |
ENST00000371817.3
|
COL5A1
|
collagen, type V, alpha 1 |
| chr19_-_1401486 | 4.03 |
ENST00000252288.2
ENST00000447102.3 |
GAMT
|
guanidinoacetate N-methyltransferase |
| chr17_+_42264556 | 4.02 |
ENST00000319511.6
ENST00000589785.1 ENST00000592825.1 ENST00000589184.1 |
TMUB2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr2_-_242255117 | 4.02 |
ENST00000420451.1
ENST00000417540.1 ENST00000310931.4 |
HDLBP
|
high density lipoprotein binding protein |
| chr19_+_50353944 | 3.99 |
ENST00000594151.1
ENST00000600603.1 ENST00000601638.1 ENST00000221557.9 |
PTOV1
|
prostate tumor overexpressed 1 |
| chr17_+_58227287 | 3.98 |
ENST00000300900.4
ENST00000591725.1 |
CA4
|
carbonic anhydrase IV |
| chr17_+_7211656 | 3.95 |
ENST00000416016.2
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr22_-_44258360 | 3.93 |
ENST00000330884.4
ENST00000249130.5 |
SULT4A1
|
sulfotransferase family 4A, member 1 |
| chrX_+_135229559 | 3.92 |
ENST00000394155.2
|
FHL1
|
four and a half LIM domains 1 |
| chrX_+_135229731 | 3.91 |
ENST00000420362.1
|
FHL1
|
four and a half LIM domains 1 |
| chr8_+_26371763 | 3.89 |
ENST00000521913.1
|
DPYSL2
|
dihydropyrimidinase-like 2 |
| chr12_+_12878829 | 3.87 |
ENST00000326765.6
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr11_-_46940074 | 3.78 |
ENST00000378623.1
ENST00000534404.1 |
LRP4
|
low density lipoprotein receptor-related protein 4 |
| chr15_-_71146480 | 3.78 |
ENST00000299213.8
|
LARP6
|
La ribonucleoprotein domain family, member 6 |
| chr20_-_524362 | 3.74 |
ENST00000460062.2
ENST00000608066.1 |
CSNK2A1
|
casein kinase 2, alpha 1 polypeptide |
| chr20_+_44657845 | 3.73 |
ENST00000243964.3
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5 |
| chr11_+_64067809 | 3.72 |
ENST00000328404.6
ENST00000539943.1 |
TEX40
|
testis expressed 40 |
| chr6_-_34664612 | 3.61 |
ENST00000374023.3
ENST00000374026.3 |
C6orf106
|
chromosome 6 open reading frame 106 |
| chr17_+_59477233 | 3.59 |
ENST00000240328.3
|
TBX2
|
T-box 2 |
| chr17_+_40811283 | 3.58 |
ENST00000251412.7
|
TUBG2
|
tubulin, gamma 2 |
| chr15_+_74218787 | 3.57 |
ENST00000261921.7
|
LOXL1
|
lysyl oxidase-like 1 |
| chr2_+_113816685 | 3.57 |
ENST00000393200.2
|
IL36RN
|
interleukin 36 receptor antagonist |
| chr12_+_117176090 | 3.55 |
ENST00000257575.4
ENST00000407967.3 ENST00000392549.2 |
RNFT2
|
ring finger protein, transmembrane 2 |
| chr19_-_49658641 | 3.46 |
ENST00000252825.4
|
HRC
|
histidine rich calcium binding protein |
| chr19_+_17186577 | 3.41 |
ENST00000595618.1
ENST00000594824.1 |
MYO9B
|
myosin IXB |
| chr5_+_170288856 | 3.40 |
ENST00000523189.1
|
RANBP17
|
RAN binding protein 17 |
| chr1_+_180601139 | 3.38 |
ENST00000367590.4
ENST00000367589.3 |
XPR1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr19_+_56186557 | 3.36 |
ENST00000270460.6
|
EPN1
|
epsin 1 |
| chr6_+_43737939 | 3.31 |
ENST00000372067.3
|
VEGFA
|
vascular endothelial growth factor A |
| chr16_+_2039946 | 3.31 |
ENST00000248121.2
ENST00000568896.1 |
SYNGR3
|
synaptogyrin 3 |
| chr11_-_46722117 | 3.30 |
ENST00000311956.4
|
ARHGAP1
|
Rho GTPase activating protein 1 |
| chr19_+_56186606 | 3.27 |
ENST00000085079.7
|
EPN1
|
epsin 1 |
| chr8_+_27631903 | 3.26 |
ENST00000305188.8
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr6_-_132272504 | 3.24 |
ENST00000367976.3
|
CTGF
|
connective tissue growth factor |
| chr11_+_61891445 | 3.24 |
ENST00000394818.3
ENST00000533896.1 ENST00000278849.4 |
INCENP
|
inner centromere protein antigens 135/155kDa |
| chr8_-_26371608 | 3.22 |
ENST00000522362.2
|
PNMA2
|
paraneoplastic Ma antigen 2 |
| chr19_-_49622348 | 3.22 |
ENST00000408991.2
|
C19orf73
|
chromosome 19 open reading frame 73 |
| chr18_+_56530794 | 3.19 |
ENST00000590285.1
ENST00000586085.1 ENST00000589288.1 |
ZNF532
|
zinc finger protein 532 |
| chr6_+_42018251 | 3.19 |
ENST00000372978.3
ENST00000494547.1 ENST00000456846.2 ENST00000372982.4 ENST00000472818.1 ENST00000372977.3 |
TAF8
|
TAF8 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 43kDa |
| chr19_-_56632592 | 3.18 |
ENST00000587279.1
ENST00000270459.3 |
ZNF787
|
zinc finger protein 787 |
| chr5_+_80529104 | 3.17 |
ENST00000254035.4
ENST00000511719.1 ENST00000437669.1 ENST00000424301.2 ENST00000505060.1 |
CKMT2
|
creatine kinase, mitochondrial 2 (sarcomeric) |
| chr5_-_131562935 | 3.16 |
ENST00000379104.2
ENST00000379100.2 ENST00000428369.1 |
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II |
| chr3_+_14989076 | 3.13 |
ENST00000413118.1
ENST00000425241.1 |
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr16_-_30582888 | 3.13 |
ENST00000563707.1
ENST00000567855.1 |
ZNF688
|
zinc finger protein 688 |
| chr10_+_49514698 | 3.10 |
ENST00000432379.1
ENST00000429041.1 ENST00000374189.1 |
MAPK8
|
mitogen-activated protein kinase 8 |
| chr4_+_20255123 | 3.10 |
ENST00000504154.1
ENST00000273739.5 |
SLIT2
|
slit homolog 2 (Drosophila) |
| chrX_-_47509887 | 3.09 |
ENST00000247161.3
ENST00000592066.1 ENST00000376983.3 |
ELK1
|
ELK1, member of ETS oncogene family |
| chr19_+_50432400 | 3.07 |
ENST00000423777.2
ENST00000600336.1 ENST00000597227.1 |
ATF5
|
activating transcription factor 5 |
| chr19_-_14117074 | 3.07 |
ENST00000588885.1
ENST00000254325.4 |
RFX1
|
regulatory factor X, 1 (influences HLA class II expression) |
| chr13_+_37006421 | 3.05 |
ENST00000255465.4
|
CCNA1
|
cyclin A1 |
| chr22_-_36903069 | 3.04 |
ENST00000216187.6
ENST00000423980.1 |
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr17_+_42923686 | 3.03 |
ENST00000591513.1
|
HIGD1B
|
HIG1 hypoxia inducible domain family, member 1B |
| chr17_+_7184986 | 3.02 |
ENST00000317370.8
ENST00000571308.1 |
SLC2A4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr11_+_62439126 | 2.99 |
ENST00000377953.3
|
C11orf83
|
chromosome 11 open reading frame 83 |
| chr6_+_127587755 | 2.97 |
ENST00000368314.1
ENST00000476956.1 ENST00000609447.1 ENST00000356799.2 ENST00000477776.1 ENST00000609944.1 |
RNF146
|
ring finger protein 146 |
| chr20_-_50808236 | 2.97 |
ENST00000361387.2
|
ZFP64
|
ZFP64 zinc finger protein |
| chr19_-_49658387 | 2.96 |
ENST00000595625.1
|
HRC
|
histidine rich calcium binding protein |
| chr7_+_100318423 | 2.95 |
ENST00000252723.2
|
EPO
|
erythropoietin |
| chr13_+_37006398 | 2.94 |
ENST00000418263.1
|
CCNA1
|
cyclin A1 |
| chr17_-_73511584 | 2.94 |
ENST00000321617.3
|
CASKIN2
|
CASK interacting protein 2 |
| chr11_+_46722368 | 2.92 |
ENST00000311764.2
|
ZNF408
|
zinc finger protein 408 |
| chr16_+_67880820 | 2.89 |
ENST00000567105.1
|
NUTF2
|
nuclear transport factor 2 |
| chr6_+_43738444 | 2.88 |
ENST00000324450.6
ENST00000417285.2 ENST00000413642.3 ENST00000372055.4 ENST00000482630.2 ENST00000425836.2 ENST00000372064.4 ENST00000372077.4 ENST00000519767.1 |
VEGFA
|
vascular endothelial growth factor A |
| chr2_+_54342574 | 2.87 |
ENST00000303536.4
ENST00000394666.3 |
ACYP2
|
acylphosphatase 2, muscle type |
| chr1_-_154946825 | 2.87 |
ENST00000368453.4
ENST00000368450.1 ENST00000366442.2 |
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr19_+_50706866 | 2.84 |
ENST00000440075.2
ENST00000376970.2 ENST00000425460.1 ENST00000599920.1 ENST00000601313.1 |
MYH14
|
myosin, heavy chain 14, non-muscle |
| chr11_+_124609823 | 2.83 |
ENST00000412681.2
|
NRGN
|
neurogranin (protein kinase C substrate, RC3) |
| chr3_+_14989186 | 2.83 |
ENST00000435454.1
ENST00000323373.6 |
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr17_-_43045439 | 2.81 |
ENST00000253407.3
|
C1QL1
|
complement component 1, q subcomponent-like 1 |
| chr22_+_51176624 | 2.80 |
ENST00000216139.5
ENST00000529621.1 |
ACR
|
acrosin |
| chr7_+_94536898 | 2.78 |
ENST00000433360.1
ENST00000340694.4 ENST00000424654.1 |
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A |
| chr10_+_88718314 | 2.77 |
ENST00000348795.4
|
SNCG
|
synuclein, gamma (breast cancer-specific protein 1) |
| chrY_-_15591485 | 2.77 |
ENST00000382896.4
ENST00000537580.1 ENST00000540140.1 ENST00000545955.1 ENST00000538878.1 |
UTY
|
ubiquitously transcribed tetratricopeptide repeat containing, Y-linked |
| chr19_-_6279932 | 2.77 |
ENST00000252674.7
|
MLLT1
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 1 |
| chr2_+_231729615 | 2.76 |
ENST00000326427.6
ENST00000335005.6 ENST00000326407.6 |
ITM2C
|
integral membrane protein 2C |
| chr19_+_49622646 | 2.74 |
ENST00000334186.4
|
PPFIA3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr6_-_167797887 | 2.73 |
ENST00000476779.2
ENST00000460930.2 ENST00000397829.4 ENST00000366827.2 |
TCP10
|
t-complex 10 |
| chr1_+_36023370 | 2.71 |
ENST00000356090.4
ENST00000373243.2 |
NCDN
|
neurochondrin |
| chr20_+_62694461 | 2.70 |
ENST00000343484.5
ENST00000395053.3 |
TCEA2
|
transcription elongation factor A (SII), 2 |
| chr8_-_42397037 | 2.70 |
ENST00000342228.3
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr3_-_178789993 | 2.69 |
ENST00000432729.1
|
ZMAT3
|
zinc finger, matrin-type 3 |
| chr17_+_7210852 | 2.68 |
ENST00000576930.1
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr2_+_113763031 | 2.67 |
ENST00000259211.6
|
IL36A
|
interleukin 36, alpha |
| chr9_+_103235365 | 2.65 |
ENST00000374879.4
|
TMEFF1
|
transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr3_-_178790057 | 2.65 |
ENST00000311417.2
|
ZMAT3
|
zinc finger, matrin-type 3 |
| chr11_+_10326612 | 2.64 |
ENST00000534464.1
ENST00000530439.1 ENST00000524948.1 ENST00000528655.1 ENST00000526492.1 ENST00000525063.1 |
ADM
|
adrenomedullin |
| chr10_+_88718397 | 2.64 |
ENST00000372017.3
|
SNCG
|
synuclein, gamma (breast cancer-specific protein 1) |
| chr2_+_27301435 | 2.63 |
ENST00000380320.4
|
EMILIN1
|
elastin microfibril interfacer 1 |
| chr9_-_13279406 | 2.63 |
ENST00000546205.1
|
MPDZ
|
multiple PDZ domain protein |
| chr5_+_173472607 | 2.62 |
ENST00000303177.3
ENST00000519867.1 |
NSG2
|
Neuron-specific protein family member 2 |
| chr11_+_66036004 | 2.59 |
ENST00000311481.6
ENST00000527397.1 |
RAB1B
|
RAB1B, member RAS oncogene family |
| chr19_+_35630344 | 2.58 |
ENST00000455515.2
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr14_-_101036119 | 2.57 |
ENST00000355173.2
|
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr6_+_117002339 | 2.55 |
ENST00000413340.1
ENST00000368564.1 ENST00000356348.1 |
KPNA5
|
karyopherin alpha 5 (importin alpha 6) |
| chr22_-_51016433 | 2.54 |
ENST00000405237.3
|
CPT1B
|
carnitine palmitoyltransferase 1B (muscle) |
| chr5_+_149569520 | 2.54 |
ENST00000230671.2
ENST00000524041.1 |
SLC6A7
|
solute carrier family 6 (neurotransmitter transporter), member 7 |
| chr12_+_111471828 | 2.54 |
ENST00000261726.6
|
CUX2
|
cut-like homeobox 2 |
| chr4_+_4388805 | 2.52 |
ENST00000504171.1
|
NSG1
|
Homo sapiens neuron specific gene family member 1 (NSG1), transcript variant 3, mRNA. |
| chr4_+_128886424 | 2.52 |
ENST00000398965.1
|
C4orf29
|
chromosome 4 open reading frame 29 |
| chr1_-_46769261 | 2.52 |
ENST00000343304.6
|
LRRC41
|
leucine rich repeat containing 41 |
| chr6_-_33267101 | 2.51 |
ENST00000497454.1
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chrX_+_152990302 | 2.50 |
ENST00000218104.3
|
ABCD1
|
ATP-binding cassette, sub-family D (ALD), member 1 |
| chr4_+_40058411 | 2.48 |
ENST00000261435.6
ENST00000515550.1 |
N4BP2
|
NEDD4 binding protein 2 |
| chr19_-_2702681 | 2.48 |
ENST00000382159.3
|
GNG7
|
guanine nucleotide binding protein (G protein), gamma 7 |
| chr8_+_22423219 | 2.48 |
ENST00000523965.1
ENST00000521554.1 |
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr18_-_51750948 | 2.48 |
ENST00000583046.1
ENST00000398398.2 |
MBD2
|
methyl-CpG binding domain protein 2 |
| chr11_+_450255 | 2.48 |
ENST00000308020.5
|
PTDSS2
|
phosphatidylserine synthase 2 |
| chr21_-_47648665 | 2.47 |
ENST00000450351.1
ENST00000522411.1 ENST00000356396.4 ENST00000397728.3 ENST00000457828.2 |
LSS
|
lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) |
| chrX_+_30671476 | 2.45 |
ENST00000378946.3
ENST00000378943.3 ENST00000378945.3 ENST00000427190.1 ENST00000378941.3 |
GK
|
glycerol kinase |
| chr7_-_767249 | 2.43 |
ENST00000403562.1
|
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr7_-_766879 | 2.43 |
ENST00000537384.1
ENST00000417852.1 |
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr14_-_24551195 | 2.43 |
ENST00000560550.1
|
NRL
|
neural retina leucine zipper |
| chr16_-_28937027 | 2.43 |
ENST00000358201.4
|
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr19_+_4304632 | 2.43 |
ENST00000597590.1
|
FSD1
|
fibronectin type III and SPRY domain containing 1 |
| chr2_-_97304105 | 2.41 |
ENST00000599854.1
ENST00000441706.2 |
KANSL3
|
KAT8 regulatory NSL complex subunit 3 |
| chr4_+_668348 | 2.40 |
ENST00000511290.1
|
MYL5
|
myosin, light chain 5, regulatory |
| chr2_-_97304009 | 2.40 |
ENST00000431828.1
ENST00000435669.1 ENST00000440133.1 ENST00000448075.1 |
KANSL3
|
KAT8 regulatory NSL complex subunit 3 |
| chr9_+_37650945 | 2.38 |
ENST00000377765.3
|
FRMPD1
|
FERM and PDZ domain containing 1 |
| chr14_+_37126765 | 2.37 |
ENST00000402703.2
|
PAX9
|
paired box 9 |
| chr19_+_35759824 | 2.36 |
ENST00000343550.5
|
USF2
|
upstream transcription factor 2, c-fos interacting |
| chr19_+_48867652 | 2.35 |
ENST00000344846.2
|
SYNGR4
|
synaptogyrin 4 |
| chr3_-_48470838 | 2.34 |
ENST00000358459.4
ENST00000358536.4 |
PLXNB1
|
plexin B1 |
| chr19_+_3359561 | 2.34 |
ENST00000589123.1
ENST00000346156.5 ENST00000395111.3 ENST00000586919.1 |
NFIC
|
nuclear factor I/C (CCAAT-binding transcription factor) |
| chr20_+_62694590 | 2.33 |
ENST00000339217.4
|
TCEA2
|
transcription elongation factor A (SII), 2 |
| chr9_+_34179003 | 2.33 |
ENST00000545103.1
ENST00000543944.1 ENST00000536252.1 ENST00000540348.1 ENST00000297661.4 ENST00000379186.4 |
UBAP1
|
ubiquitin associated protein 1 |
| chr19_-_43708378 | 2.33 |
ENST00000599746.1
|
PSG4
|
pregnancy specific beta-1-glycoprotein 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of RXRG
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 13.6 | GO:2000777 | positive regulation of oocyte maturation(GO:1900195) negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 3.1 | 9.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 2.7 | 8.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 2.5 | 15.0 | GO:0010732 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 2.1 | 6.4 | GO:1901899 | positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 2.1 | 16.6 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 2.1 | 6.2 | GO:1903572 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 1.9 | 7.4 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 1.8 | 5.4 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 1.5 | 4.6 | GO:0006258 | UDP-glucose catabolic process(GO:0006258) |
| 1.4 | 4.3 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 1.4 | 4.2 | GO:1900104 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 1.4 | 8.4 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 1.4 | 4.2 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 1.3 | 3.8 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 1.3 | 5.0 | GO:1904046 | negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 1.2 | 4.8 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 1.2 | 4.6 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 1.1 | 4.6 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 1.1 | 1.1 | GO:1990641 | response to iron ion starvation(GO:1990641) |
| 1.1 | 4.5 | GO:0061304 | extracellular matrix-cell signaling(GO:0035426) retinal blood vessel morphogenesis(GO:0061304) |
| 1.1 | 3.3 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 1.1 | 3.2 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 1.1 | 4.3 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 1.1 | 4.3 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 1.0 | 5.2 | GO:0072240 | thick ascending limb development(GO:0072023) DCT cell differentiation(GO:0072069) metanephric thick ascending limb development(GO:0072233) metanephric DCT cell differentiation(GO:0072240) |
| 1.0 | 3.1 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 1.0 | 3.1 | GO:0021966 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) corticospinal neuron axon guidance(GO:0021966) regulation of negative chemotaxis(GO:0050923) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 1.0 | 9.1 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 1.0 | 4.0 | GO:0019086 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) late viral transcription(GO:0019086) |
| 1.0 | 2.9 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 1.0 | 5.8 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.9 | 2.8 | GO:0061743 | motor learning(GO:0061743) |
| 0.9 | 3.7 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.9 | 2.7 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.9 | 2.7 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.9 | 4.3 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.8 | 2.5 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.8 | 2.3 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.7 | 4.4 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.7 | 7.2 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.7 | 3.6 | GO:0007521 | muscle cell fate determination(GO:0007521) |
| 0.7 | 2.8 | GO:0033123 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.7 | 2.1 | GO:0090298 | negative regulation of mitochondrial DNA replication(GO:0090298) negative regulation of mitochondrial DNA metabolic process(GO:1901859) |
| 0.7 | 5.6 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.7 | 2.8 | GO:1904048 | regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.7 | 2.7 | GO:0002884 | regulation of type IV hypersensitivity(GO:0001807) negative regulation of hypersensitivity(GO:0002884) |
| 0.7 | 3.4 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.7 | 3.4 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.7 | 2.0 | GO:0003094 | glomerular filtration(GO:0003094) renal filtration(GO:0097205) |
| 0.7 | 2.0 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) cell migration involved in heart formation(GO:0060974) anterior neural tube closure(GO:0061713) |
| 0.7 | 3.3 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.7 | 3.9 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.6 | 6.3 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.6 | 6.3 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.6 | 2.5 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.6 | 2.4 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.6 | 3.6 | GO:1902714 | negative regulation of interferon-gamma secretion(GO:1902714) |
| 0.6 | 4.7 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.6 | 5.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.5 | 3.7 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.5 | 3.1 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.5 | 2.5 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.5 | 9.0 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.5 | 2.9 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.5 | 1.9 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) regulation of telomeric DNA binding(GO:1904742) |
| 0.5 | 1.4 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.5 | 0.9 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.5 | 3.3 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.5 | 1.8 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.5 | 5.5 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.5 | 1.8 | GO:0035962 | response to interleukin-13(GO:0035962) |
| 0.4 | 1.3 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.4 | 1.8 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.4 | 4.6 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.4 | 1.2 | GO:1901631 | positive regulation of presynaptic membrane organization(GO:1901631) |
| 0.4 | 1.2 | GO:0021637 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.4 | 5.4 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.4 | 1.5 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.4 | 1.9 | GO:1901569 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.4 | 1.8 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.4 | 1.8 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.4 | 1.5 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.4 | 1.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.4 | 3.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.4 | 1.8 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.4 | 1.8 | GO:0010868 | negative regulation of triglyceride biosynthetic process(GO:0010868) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.4 | 5.3 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.3 | 3.8 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.3 | 4.5 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.3 | 2.8 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.3 | 1.7 | GO:0070384 | growth plate cartilage chondrocyte growth(GO:0003430) growth plate cartilage chondrocyte development(GO:0003431) Harderian gland development(GO:0070384) |
| 0.3 | 2.7 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.3 | 7.9 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.3 | 2.0 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.3 | 1.6 | GO:0001834 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.3 | 1.6 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.3 | 1.0 | GO:1902617 | response to fluoride(GO:1902617) |
| 0.3 | 2.5 | GO:0099639 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.3 | 3.1 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.3 | 4.9 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.3 | 4.8 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.3 | 1.8 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.3 | 1.8 | GO:0033504 | floor plate development(GO:0033504) |
| 0.3 | 2.1 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.3 | 1.2 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.3 | 4.0 | GO:0006705 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.3 | 2.3 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.3 | 5.1 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.3 | 2.8 | GO:0021853 | cerebral cortex tangential migration(GO:0021800) cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.3 | 1.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.3 | 0.3 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.3 | 2.2 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.3 | 1.3 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.3 | 1.9 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.3 | 4.0 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.3 | 1.1 | GO:0009227 | nucleotide-sugar catabolic process(GO:0009227) |
| 0.3 | 3.7 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.3 | 0.8 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.3 | 2.3 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.2 | 2.0 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.2 | 0.7 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.2 | 1.9 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.2 | 1.2 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.2 | 1.0 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.2 | 1.4 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.2 | 2.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.2 | 3.7 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.2 | 2.7 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 | 2.3 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to photoreceptor outer segment(GO:1903546) |
| 0.2 | 1.4 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.2 | 12.0 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.2 | 0.7 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.2 | 0.8 | GO:0032972 | diaphragm contraction(GO:0002086) regulation of muscle filament sliding speed(GO:0032972) |
| 0.2 | 2.1 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 0.8 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.2 | 1.9 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.2 | 1.7 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.2 | 6.8 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.2 | 2.9 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.2 | 1.8 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.2 | 1.2 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.2 | 2.8 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.2 | 0.8 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.2 | 3.6 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.2 | 0.6 | GO:0072093 | negative regulation of hair follicle development(GO:0051799) metanephric renal vesicle formation(GO:0072093) |
| 0.2 | 18.1 | GO:0005977 | glycogen metabolic process(GO:0005977) |
| 0.2 | 1.8 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.2 | 1.4 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.2 | 2.5 | GO:0007614 | short-term memory(GO:0007614) |
| 0.2 | 1.8 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.2 | 0.2 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.2 | 1.1 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.2 | 1.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.2 | 0.8 | GO:0036114 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.2 | 2.3 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.2 | 1.7 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.2 | 1.9 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.2 | 1.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.2 | 2.7 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.2 | 0.9 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.2 | 4.7 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.2 | 1.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.2 | 1.6 | GO:0035376 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.2 | 0.5 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.2 | 1.4 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.2 | 0.5 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.2 | 1.5 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.2 | 4.1 | GO:0050427 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.2 | 4.1 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.2 | 0.5 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.2 | 1.8 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.2 | 2.6 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.2 | 2.8 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.2 | 0.5 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.2 | 0.6 | GO:0021784 | postganglionic parasympathetic fiber development(GO:0021784) |
| 0.2 | 2.7 | GO:0036503 | ERAD pathway(GO:0036503) |
| 0.2 | 0.8 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.2 | 0.8 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.1 | 2.5 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.1 | 4.7 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 1.7 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 4.0 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.1 | 0.9 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 5.3 | GO:0042417 | dopamine metabolic process(GO:0042417) |
| 0.1 | 1.9 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 1.8 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.1 | 7.5 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 1.0 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.1 | 2.3 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 6.1 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.1 | 0.3 | GO:1990262 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.1 | 0.9 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 0.1 | 2.6 | GO:0019068 | virion assembly(GO:0019068) |
| 0.1 | 1.5 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 3.6 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 1.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 3.0 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.1 | 6.9 | GO:0090102 | cochlea development(GO:0090102) |
| 0.1 | 2.7 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.1 | 2.8 | GO:0032607 | interferon-alpha production(GO:0032607) |
| 0.1 | 0.6 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 1.3 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.1 | 0.6 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.1 | 1.8 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 6.6 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 1.4 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.1 | 1.3 | GO:0006665 | sphingolipid metabolic process(GO:0006665) |
| 0.1 | 0.7 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.1 | 2.1 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.7 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 5.0 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.1 | 0.7 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.1 | 0.4 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.1 | 4.1 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 1.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 5.1 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.1 | 2.7 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.1 | 1.3 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.1 | 1.0 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.1 | 1.3 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.1 | 2.0 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.1 | 2.5 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.1 | 2.2 | GO:0032402 | melanosome transport(GO:0032402) |
| 0.1 | 2.2 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 2.4 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.1 | 0.8 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.1 | 1.3 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.1 | 0.9 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.5 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 0.2 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.1 | 0.3 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 | 0.6 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 1.6 | GO:0040020 | regulation of meiotic nuclear division(GO:0040020) |
| 0.1 | 2.5 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.1 | 1.9 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 1.4 | GO:1900273 | positive regulation of long-term synaptic potentiation(GO:1900273) |
| 0.1 | 0.5 | GO:0033692 | cellular polysaccharide biosynthetic process(GO:0033692) |
| 0.1 | 0.9 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 0.2 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.1 | 4.0 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 0.9 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.1 | 4.4 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 3.3 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.1 | 1.9 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.1 | 2.8 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.1 | 1.6 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.1 | 1.2 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.1 | 1.0 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 2.3 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 1.8 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 0.9 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 0.9 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.1 | 5.7 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.1 | 0.6 | GO:0071168 | protein localization to chromatin(GO:0071168) |
| 0.1 | 0.6 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.1 | 0.2 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.1 | 1.6 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 1.5 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 2.0 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 1.5 | GO:0051438 | regulation of ubiquitin-protein transferase activity(GO:0051438) |
| 0.1 | 1.1 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.1 | 5.9 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.1 | 0.5 | GO:0045019 | negative regulation of nitric oxide biosynthetic process(GO:0045019) negative regulation of nitric oxide metabolic process(GO:1904406) |
| 0.1 | 2.0 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 0.3 | GO:0030047 | actin modification(GO:0030047) |
| 0.1 | 0.2 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.1 | 1.9 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 2.0 | GO:0002027 | regulation of heart rate(GO:0002027) |
| 0.0 | 1.3 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 0.2 | GO:0032594 | protein transport within lipid bilayer(GO:0032594) protein transport into membrane raft(GO:0032596) |
| 0.0 | 1.1 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.9 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 1.9 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 1.5 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 1.5 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 1.7 | GO:0051058 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 0.4 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 2.5 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 1.6 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.0 | 0.9 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 2.5 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.8 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.2 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 1.7 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.1 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.5 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 1.0 | GO:0048536 | spleen development(GO:0048536) |
| 0.0 | 0.4 | GO:0018022 | peptidyl-lysine methylation(GO:0018022) |
| 0.0 | 2.7 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 1.5 | GO:0007565 | female pregnancy(GO:0007565) |
| 0.0 | 1.8 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 1.2 | GO:0035418 | protein localization to synapse(GO:0035418) |
| 0.0 | 0.9 | GO:0009083 | branched-chain amino acid metabolic process(GO:0009081) branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.3 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.2 | GO:0007512 | adult heart development(GO:0007512) |
| 0.0 | 1.5 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.0 | 1.1 | GO:0006040 | amino sugar metabolic process(GO:0006040) |
| 0.0 | 1.5 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.7 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 1.2 | GO:0007017 | microtubule-based process(GO:0007017) |
| 0.0 | 2.8 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.5 | GO:0039694 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 1.3 | GO:0007188 | adenylate cyclase-modulating G-protein coupled receptor signaling pathway(GO:0007188) |
| 0.0 | 1.2 | GO:0001764 | neuron migration(GO:0001764) |
| 0.0 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.5 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 0.1 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 1.1 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 3.5 | GO:0007009 | plasma membrane organization(GO:0007009) |
| 0.0 | 1.4 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.3 | GO:0006939 | smooth muscle contraction(GO:0006939) |
| 0.0 | 0.6 | GO:0030038 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.8 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.6 | GO:0042116 | macrophage activation(GO:0042116) |
| 0.0 | 0.1 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 1.1 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.9 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 2.1 | GO:0007411 | axon guidance(GO:0007411) |
| 0.0 | 0.4 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 1.8 | GO:0006986 | response to unfolded protein(GO:0006986) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 7.9 | GO:0097123 | cyclin A1-CDK2 complex(GO:0097123) |
| 1.5 | 13.6 | GO:0072687 | meiotic spindle(GO:0072687) |
| 1.4 | 4.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 1.1 | 2.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 1.0 | 5.8 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.9 | 2.8 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.8 | 9.8 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.8 | 19.0 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.8 | 15.0 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.7 | 6.5 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.7 | 6.4 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.7 | 7.5 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.7 | 4.6 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.6 | 6.5 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.6 | 3.2 | GO:0000801 | central element(GO:0000801) |
| 0.6 | 5.1 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.6 | 12.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.6 | 2.8 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.6 | 2.3 | GO:0043293 | apoptosome(GO:0043293) |
| 0.6 | 2.8 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.5 | 3.3 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.5 | 1.5 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.5 | 2.9 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.5 | 6.4 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.4 | 6.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.4 | 2.5 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.4 | 1.7 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.4 | 3.7 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.4 | 6.0 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.4 | 5.0 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.3 | 1.4 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.3 | 2.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.3 | 3.7 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.3 | 2.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.3 | 3.6 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.3 | 2.5 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 0.3 | 4.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.3 | 4.4 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.3 | 2.8 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.3 | 1.9 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.3 | 4.9 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.3 | 1.1 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.3 | 1.9 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.3 | 1.8 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.2 | 3.3 | GO:0001741 | XY body(GO:0001741) |
| 0.2 | 1.8 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.2 | 1.1 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.2 | 2.8 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.2 | 1.7 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 2.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 1.8 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 7.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 1.6 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.2 | 3.1 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.2 | 6.2 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.2 | 11.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.2 | 2.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.2 | 14.3 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.2 | 1.5 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.2 | 4.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.2 | 1.5 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.2 | 1.2 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 2.0 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.6 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 2.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 5.2 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 0.8 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 0.9 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 5.0 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.1 | 6.9 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 2.6 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.1 | 1.5 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 1.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 10.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 1.0 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 5.5 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 9.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 1.7 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 2.6 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 1.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 0.6 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.1 | 1.8 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.1 | 1.8 | GO:0042599 | lamellar body(GO:0042599) |
| 0.1 | 4.0 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 3.0 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 8.2 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 1.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 3.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 1.7 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 1.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.3 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 0.9 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.1 | 1.1 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.1 | 5.3 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 2.7 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 1.0 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.7 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.8 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 2.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 6.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 4.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 3.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 4.3 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.1 | 3.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 2.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 1.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 1.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 1.5 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 0.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 2.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 4.0 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.5 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 1.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 2.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 4.9 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 1.2 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 1.4 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.9 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 3.3 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 6.1 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 48.8 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.9 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.9 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 2.2 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 2.9 | GO:0042175 | nuclear outer membrane-endoplasmic reticulum membrane network(GO:0042175) |
| 0.0 | 0.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.6 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 2.4 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 1.6 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 3.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 5.8 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 1.0 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.9 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 1.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.5 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 1.1 | GO:0005929 | cilium(GO:0005929) |
| 0.0 | 1.1 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.4 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 3.0 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.9 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.7 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 3.8 | GO:0005667 | transcription factor complex(GO:0005667) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 13.6 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 2.7 | 8.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 2.3 | 6.9 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 2.2 | 17.8 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 2.1 | 8.5 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 1.9 | 5.8 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 1.7 | 6.7 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 1.6 | 4.7 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 1.4 | 4.3 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 1.4 | 1.4 | GO:0009055 | electron carrier activity(GO:0009055) |
| 1.3 | 3.9 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 1.1 | 5.3 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 1.0 | 3.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 1.0 | 6.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 1.0 | 4.0 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.9 | 2.8 | GO:0004040 | amidase activity(GO:0004040) fucose binding(GO:0042806) |
| 0.9 | 2.6 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.8 | 3.4 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.8 | 4.7 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.7 | 4.4 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.7 | 13.8 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.7 | 6.5 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.7 | 3.6 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.7 | 5.6 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.7 | 2.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) steroid delta-isomerase activity(GO:0004769) |
| 0.7 | 4.8 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.7 | 2.7 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.7 | 6.0 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.7 | 2.0 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.7 | 4.6 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.7 | 3.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.6 | 1.9 | GO:0097258 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.6 | 1.9 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.6 | 2.5 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.6 | 1.2 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.6 | 1.7 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.5 | 3.8 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.5 | 2.6 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.5 | 2.5 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.5 | 2.5 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.5 | 2.5 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.5 | 2.0 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.5 | 3.9 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.5 | 2.9 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.5 | 1.8 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.5 | 1.8 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.5 | 7.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.4 | 1.3 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.4 | 3.6 | GO:0016679 | oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) |
| 0.4 | 17.9 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.4 | 4.6 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.4 | 2.9 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.4 | 3.3 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.4 | 2.0 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.4 | 3.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.4 | 8.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.4 | 1.5 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.4 | 2.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.4 | 9.4 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.4 | 2.5 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.3 | 1.4 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.3 | 1.4 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.3 | 3.7 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.3 | 1.7 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.3 | 2.7 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.3 | 1.3 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.3 | 4.5 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.3 | 4.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.3 | 2.5 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.3 | 1.9 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.3 | 1.5 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.3 | 1.5 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.3 | 1.8 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.3 | 1.4 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.3 | 3.4 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.3 | 0.8 | GO:0052830 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol tetrakisphosphate kinase activity(GO:0051765) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) |
| 0.3 | 1.1 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.3 | 4.8 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.2 | 1.7 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.2 | 1.0 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.2 | 2.8 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.2 | 4.0 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 5.7 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.2 | 5.2 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.2 | 3.0 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.2 | 2.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.2 | 1.5 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.2 | 1.7 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.2 | 3.6 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.2 | 4.4 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.2 | 1.7 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.2 | 10.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 2.8 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.2 | 0.8 | GO:0004566 | beta-glucuronidase activity(GO:0004566) beta-glucosidase activity(GO:0008422) |
| 0.2 | 3.6 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.2 | 9.5 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.2 | 1.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.2 | 1.6 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.2 | 1.6 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.2 | 1.6 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.2 | 2.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.2 | 3.9 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.2 | 3.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 4.7 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.2 | 2.7 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.2 | 0.2 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.2 | 1.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.2 | 1.3 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.2 | 3.1 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.2 | 5.0 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.2 | 1.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 0.9 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.9 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 1.0 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 1.1 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.1 | 21.4 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 1.8 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 0.4 | GO:0080101 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.1 | 4.7 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 1.7 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 3.9 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 1.9 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 1.2 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 1.1 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.1 | 1.0 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 6.2 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 2.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 1.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 0.5 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 1.5 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 2.2 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 14.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 5.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 1.7 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 11.4 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 2.5 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 1.2 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 0.3 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) oncostatin-M receptor activity(GO:0004924) interleukin-6 binding(GO:0019981) |
| 0.1 | 2.2 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 2.5 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 1.1 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.1 | 1.6 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.1 | 2.7 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.6 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 3.1 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.1 | 0.8 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 1.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 1.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 5.9 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 1.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 1.9 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.9 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 2.3 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 4.1 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 0.8 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.1 | 0.8 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 0.8 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 1.8 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 1.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.5 | GO:0098988 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) kainate selective glutamate receptor activity(GO:0015277) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.1 | 2.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 2.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 1.1 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.1 | 2.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 0.5 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.3 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.1 | 0.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 2.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 1.5 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 1.0 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 0.6 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 25.3 | GO:0005525 | GTP binding(GO:0005525) |
| 0.1 | 1.9 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 5.1 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 1.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 2.4 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 0.9 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.5 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 7.0 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 1.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.5 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 2.0 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 1.9 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 9.0 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.9 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 8.9 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 0.9 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.6 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 2.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 1.1 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.0 | 0.3 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 2.9 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 3.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 1.2 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 3.0 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) arginine binding(GO:0034618) |
| 0.0 | 0.8 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.7 | GO:0008066 | glutamate receptor activity(GO:0008066) |
| 0.0 | 1.6 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 2.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.6 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 2.8 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.9 | GO:0061733 | histone acetyltransferase activity(GO:0004402) peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.0 | 0.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 4.6 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 10.7 | GO:0008289 | lipid binding(GO:0008289) |
| 0.0 | 0.4 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 2.5 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.4 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.7 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 14.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.4 | 9.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.3 | 17.7 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.3 | 19.7 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.3 | 7.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.3 | 7.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.2 | 3.7 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.2 | 9.8 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.2 | 5.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.2 | 2.9 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 8.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 5.8 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 7.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 3.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 2.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 2.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 4.6 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 4.7 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 1.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 4.9 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 1.8 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 3.4 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.1 | 2.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 3.6 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 1.9 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 2.7 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 4.0 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 1.7 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 1.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 2.4 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 4.7 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 5.2 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 2.7 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 1.6 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 4.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 4.4 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 2.4 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.1 | 16.4 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.9 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.1 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 2.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 10.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 9.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.8 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 5.7 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 0.3 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.2 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 1.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.2 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.9 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.5 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 16.6 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.4 | 11.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.4 | 3.3 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.4 | 4.7 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.4 | 6.8 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.3 | 7.2 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.3 | 6.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.3 | 7.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.3 | 13.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.3 | 6.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.3 | 4.0 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.3 | 3.6 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.2 | 5.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 4.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 3.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.2 | 2.7 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.2 | 1.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 1.0 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.2 | 8.0 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 8.6 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.2 | 7.0 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.2 | 4.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 3.5 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.2 | 4.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.2 | 6.0 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.2 | 0.5 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.2 | 4.8 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.2 | 5.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 3.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.2 | 3.0 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 2.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 2.9 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 1.8 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 3.9 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 2.9 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 2.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 2.8 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 3.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 2.5 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 7.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 4.2 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 1.9 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 3.5 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.1 | 9.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 2.1 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.1 | 3.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 2.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 1.1 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 5.3 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.1 | 7.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 3.1 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 1.6 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.8 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 1.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 2.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 0.6 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.1 | 7.4 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.1 | 1.9 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 1.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 0.9 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 1.9 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 0.9 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 2.6 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 1.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 2.5 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.1 | 0.5 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |
| 0.1 | 1.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 0.8 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 0.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 2.5 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 7.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 3.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.5 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 0.9 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 2.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 4.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.7 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.5 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.8 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.0 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.8 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |