Project
averaged GNF SymAtlas + NCI-60 cancer cell lines, human (Su, 2004; Ross, 2000)
Navigation
Downloads
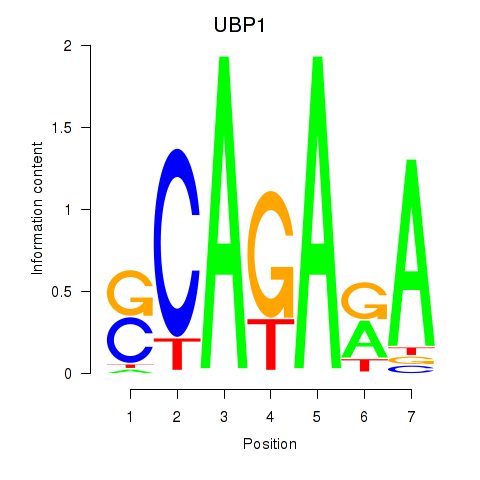
Results for UBP1
Z-value: 0.57
Transcription factors associated with UBP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
UBP1
|
ENSG00000153560.7 | upstream binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| UBP1 | hg19_v2_chr3_-_33481835_33481905 | 0.02 | 8.0e-01 | Click! |
Activity profile of UBP1 motif
Sorted Z-values of UBP1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_5138099 | 10.69 |
ENST00000571800.1
ENST00000574081.1 ENST00000399600.4 ENST00000574297.1 |
SCIMP
|
SLP adaptor and CSK interacting membrane protein |
| chr7_+_142498725 | 9.70 |
ENST00000466254.1
|
TRBC2
|
T cell receptor beta constant 2 |
| chr22_+_22707260 | 7.12 |
ENST00000390293.1
|
IGLV5-48
|
immunoglobulin lambda variable 5-48 (non-functional) |
| chr6_+_32709119 | 6.00 |
ENST00000374940.3
|
HLA-DQA2
|
major histocompatibility complex, class II, DQ alpha 2 |
| chr2_-_89442621 | 5.11 |
ENST00000492167.1
|
IGKV3-20
|
immunoglobulin kappa variable 3-20 |
| chr1_-_150208320 | 4.16 |
ENST00000534220.1
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr6_+_31555045 | 3.59 |
ENST00000396101.3
ENST00000490742.1 |
LST1
|
leukocyte specific transcript 1 |
| chr22_+_22676808 | 3.45 |
ENST00000390290.2
|
IGLV1-51
|
immunoglobulin lambda variable 1-51 |
| chr2_+_113033164 | 3.07 |
ENST00000409871.1
ENST00000343936.4 |
ZC3H6
|
zinc finger CCCH-type containing 6 |
| chr19_-_51875523 | 2.73 |
ENST00000593572.1
ENST00000595157.1 |
NKG7
|
natural killer cell group 7 sequence |
| chr1_-_202129105 | 2.72 |
ENST00000367279.4
|
PTPN7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr1_-_202129704 | 2.57 |
ENST00000476061.1
ENST00000544762.1 ENST00000467283.1 ENST00000464870.1 ENST00000435759.2 ENST00000486116.1 ENST00000543735.1 ENST00000308986.5 ENST00000477625.1 |
PTPN7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr6_+_6588316 | 2.37 |
ENST00000379953.2
|
LY86
|
lymphocyte antigen 86 |
| chr15_+_81475047 | 2.11 |
ENST00000559388.1
|
IL16
|
interleukin 16 |
| chr5_-_111093759 | 2.06 |
ENST00000509979.1
ENST00000513100.1 ENST00000508161.1 ENST00000455559.2 |
NREP
|
neuronal regeneration related protein |
| chr3_+_111260954 | 1.93 |
ENST00000283285.5
|
CD96
|
CD96 molecule |
| chr12_-_8815299 | 1.93 |
ENST00000535336.1
|
MFAP5
|
microfibrillar associated protein 5 |
| chr9_-_35685452 | 1.70 |
ENST00000607559.1
|
TPM2
|
tropomyosin 2 (beta) |
| chr12_-_8815215 | 1.70 |
ENST00000544889.1
ENST00000543369.1 |
MFAP5
|
microfibrillar associated protein 5 |
| chr3_+_111260856 | 1.61 |
ENST00000352690.4
|
CD96
|
CD96 molecule |
| chr4_-_57524061 | 1.50 |
ENST00000508121.1
|
HOPX
|
HOP homeobox |
| chr21_+_43823983 | 1.49 |
ENST00000291535.6
ENST00000450356.1 ENST00000319294.6 ENST00000398367.1 |
UBASH3A
|
ubiquitin associated and SH3 domain containing A |
| chr8_+_28747884 | 1.40 |
ENST00000287701.10
ENST00000444075.1 ENST00000403668.2 ENST00000519662.1 ENST00000558662.1 ENST00000523613.1 ENST00000560599.1 ENST00000397358.3 |
HMBOX1
|
homeobox containing 1 |
| chr19_-_54784937 | 1.32 |
ENST00000434421.1
ENST00000314446.5 ENST00000391749.4 |
LILRB2
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 2 |
| chrX_-_65253506 | 1.23 |
ENST00000427538.1
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr12_-_47473425 | 1.22 |
ENST00000550413.1
|
AMIGO2
|
adhesion molecule with Ig-like domain 2 |
| chr22_+_38071615 | 1.18 |
ENST00000215909.5
|
LGALS1
|
lectin, galactoside-binding, soluble, 1 |
| chr11_+_10326612 | 1.17 |
ENST00000534464.1
ENST00000530439.1 ENST00000524948.1 ENST00000528655.1 ENST00000526492.1 ENST00000525063.1 |
ADM
|
adrenomedullin |
| chr1_-_150669604 | 1.13 |
ENST00000427665.1
ENST00000540514.1 |
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr17_-_62493131 | 1.08 |
ENST00000539111.2
|
POLG2
|
polymerase (DNA directed), gamma 2, accessory subunit |
| chr4_+_156588115 | 1.06 |
ENST00000455639.2
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr22_+_22385332 | 1.05 |
ENST00000390282.2
|
IGLV4-69
|
immunoglobulin lambda variable 4-69 |
| chr8_-_28747424 | 1.03 |
ENST00000523436.1
ENST00000397363.4 ENST00000521777.1 ENST00000520184.1 ENST00000521022.1 |
INTS9
|
integrator complex subunit 9 |
| chr8_-_66474884 | 1.03 |
ENST00000520902.1
|
CTD-3025N20.2
|
CTD-3025N20.2 |
| chr4_+_156587853 | 1.00 |
ENST00000506455.1
ENST00000511108.1 |
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr12_+_123717458 | 0.98 |
ENST00000253233.1
|
C12orf65
|
chromosome 12 open reading frame 65 |
| chr8_-_28747717 | 0.97 |
ENST00000416984.2
|
INTS9
|
integrator complex subunit 9 |
| chr1_-_152332480 | 0.97 |
ENST00000388718.5
|
FLG2
|
filaggrin family member 2 |
| chr7_+_101460882 | 0.95 |
ENST00000292535.7
ENST00000549414.2 ENST00000550008.2 ENST00000546411.2 ENST00000556210.1 |
CUX1
|
cut-like homeobox 1 |
| chr16_-_67970990 | 0.94 |
ENST00000358514.4
|
PSMB10
|
proteasome (prosome, macropain) subunit, beta type, 10 |
| chr5_-_93447333 | 0.93 |
ENST00000395965.3
ENST00000505869.1 ENST00000509163.1 |
FAM172A
|
family with sequence similarity 172, member A |
| chr7_+_29874341 | 0.93 |
ENST00000409290.1
ENST00000242140.5 |
WIPF3
|
WAS/WASL interacting protein family, member 3 |
| chr15_-_77712477 | 0.89 |
ENST00000560626.2
|
PEAK1
|
pseudopodium-enriched atypical kinase 1 |
| chrX_+_38420623 | 0.88 |
ENST00000378482.2
|
TSPAN7
|
tetraspanin 7 |
| chr19_-_11039188 | 0.85 |
ENST00000588347.1
|
YIPF2
|
Yip1 domain family, member 2 |
| chrX_+_13587712 | 0.81 |
ENST00000361306.1
ENST00000380602.3 |
EGFL6
|
EGF-like-domain, multiple 6 |
| chr7_+_142829162 | 0.76 |
ENST00000291009.3
|
PIP
|
prolactin-induced protein |
| chr1_-_168698433 | 0.73 |
ENST00000367817.3
|
DPT
|
dermatopontin |
| chr3_+_29323043 | 0.71 |
ENST00000452462.1
ENST00000456853.1 |
RBMS3
|
RNA binding motif, single stranded interacting protein 3 |
| chr4_+_156587979 | 0.70 |
ENST00000511507.1
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr11_+_128761220 | 0.68 |
ENST00000529694.1
|
KCNJ5
|
potassium inwardly-rectifying channel, subfamily J, member 5 |
| chr14_-_92413727 | 0.63 |
ENST00000267620.10
|
FBLN5
|
fibulin 5 |
| chr4_-_168155700 | 0.63 |
ENST00000357545.4
ENST00000512648.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr8_+_144373550 | 0.59 |
ENST00000330143.3
ENST00000521537.1 ENST00000518432.1 ENST00000520333.1 |
ZNF696
|
zinc finger protein 696 |
| chr4_-_168155730 | 0.55 |
ENST00000502330.1
ENST00000357154.3 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr12_-_123717643 | 0.55 |
ENST00000541437.1
ENST00000606320.1 |
MPHOSPH9
|
M-phase phosphoprotein 9 |
| chr6_-_29395509 | 0.55 |
ENST00000377147.2
|
OR11A1
|
olfactory receptor, family 11, subfamily A, member 1 |
| chr11_-_102668879 | 0.54 |
ENST00000315274.6
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase) |
| chr1_-_150208291 | 0.52 |
ENST00000533654.1
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr9_+_15422702 | 0.44 |
ENST00000380821.3
ENST00000421710.1 |
SNAPC3
|
small nuclear RNA activating complex, polypeptide 3, 50kDa |
| chr2_+_162087577 | 0.43 |
ENST00000439442.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr22_+_22730353 | 0.41 |
ENST00000390296.2
|
IGLV5-45
|
immunoglobulin lambda variable 5-45 |
| chr17_+_32683456 | 0.40 |
ENST00000225844.2
|
CCL13
|
chemokine (C-C motif) ligand 13 |
| chrX_+_24167828 | 0.39 |
ENST00000379188.3
ENST00000419690.1 ENST00000379177.1 ENST00000304543.5 |
ZFX
|
zinc finger protein, X-linked |
| chr4_-_168155169 | 0.36 |
ENST00000534949.1
ENST00000535728.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr4_-_47839966 | 0.35 |
ENST00000273857.4
ENST00000505909.1 ENST00000502252.1 |
CORIN
|
corin, serine peptidase |
| chr1_-_150208412 | 0.34 |
ENST00000532744.1
ENST00000369114.5 ENST00000369115.2 ENST00000369116.4 |
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr4_-_76861392 | 0.34 |
ENST00000505594.1
|
NAAA
|
N-acylethanolamine acid amidase |
| chr11_+_7506713 | 0.33 |
ENST00000329293.3
ENST00000534244.1 |
OLFML1
|
olfactomedin-like 1 |
| chr12_+_9067123 | 0.33 |
ENST00000543824.1
|
PHC1
|
polyhomeotic homolog 1 (Drosophila) |
| chr17_+_74381343 | 0.32 |
ENST00000392496.3
|
SPHK1
|
sphingosine kinase 1 |
| chr11_-_113345995 | 0.31 |
ENST00000355319.2
ENST00000542616.1 |
DRD2
|
dopamine receptor D2 |
| chr1_-_114302086 | 0.30 |
ENST00000369604.1
ENST00000357783.2 |
PHTF1
|
putative homeodomain transcription factor 1 |
| chr1_-_150208498 | 0.29 |
ENST00000314136.8
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr14_-_92413353 | 0.27 |
ENST00000556154.1
|
FBLN5
|
fibulin 5 |
| chr1_-_150208363 | 0.24 |
ENST00000436748.2
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chrX_+_115567767 | 0.22 |
ENST00000371900.4
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14 |
| chrY_+_2803322 | 0.22 |
ENST00000383052.1
ENST00000155093.3 ENST00000449237.1 ENST00000443793.1 |
ZFY
|
zinc finger protein, Y-linked |
| chr9_-_6015607 | 0.13 |
ENST00000259569.5
|
RANBP6
|
RAN binding protein 6 |
| chr2_+_17721920 | 0.12 |
ENST00000295156.4
|
VSNL1
|
visinin-like 1 |
| chr4_-_70626314 | 0.11 |
ENST00000510821.1
|
SULT1B1
|
sulfotransferase family, cytosolic, 1B, member 1 |
| chrX_-_70474910 | 0.09 |
ENST00000373988.1
ENST00000373998.1 |
ZMYM3
|
zinc finger, MYM-type 3 |
| chr13_+_24844819 | 0.07 |
ENST00000399949.2
|
SPATA13
|
spermatogenesis associated 13 |
| chr3_+_111718036 | 0.06 |
ENST00000455401.2
|
TAGLN3
|
transgelin 3 |
| chr1_+_206223941 | 0.06 |
ENST00000367126.4
|
AVPR1B
|
arginine vasopressin receptor 1B |
| chr3_-_56502375 | 0.06 |
ENST00000288221.6
|
ERC2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr2_+_61244697 | 0.05 |
ENST00000401576.1
ENST00000295030.5 ENST00000414712.2 |
PEX13
|
peroxisomal biogenesis factor 13 |
| chr14_+_21214039 | 0.00 |
ENST00000326842.2
|
EDDM3A
|
epididymal protein 3A |
Network of associatons between targets according to the STRING database.
First level regulatory network of UBP1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0002370 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 0.5 | 2.8 | GO:0052565 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.3 | 1.2 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.3 | 2.4 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.2 | 1.2 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.2 | 1.3 | GO:0002774 | Fc receptor mediated inhibitory signaling pathway(GO:0002774) |
| 0.2 | 5.1 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.2 | 1.1 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.1 | 9.7 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 5.6 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 0.3 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.1 | 0.3 | GO:0044467 | glial cell-derived neurotrophic factor secretion(GO:0044467) regulation of glial cell-derived neurotrophic factor secretion(GO:1900166) positive regulation of glial cell-derived neurotrophic factor secretion(GO:1900168) |
| 0.1 | 2.0 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 0.4 | GO:0090264 | immune complex clearance(GO:0002434) immune complex clearance by monocytes and macrophages(GO:0002436) response to high density lipoprotein particle(GO:0055099) T cell extravasation(GO:0072683) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) |
| 0.1 | 10.7 | GO:0007127 | meiosis I(GO:0007127) |
| 0.1 | 2.1 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 1.1 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.1 | 3.6 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.1 | 0.9 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 1.5 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.4 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 1.5 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.1 | 0.7 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.1 | 1.5 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 1.4 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 6.0 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.1 | 8.4 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 1.2 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 | 3.6 | GO:0032945 | negative regulation of mononuclear cell proliferation(GO:0032945) negative regulation of lymphocyte proliferation(GO:0050672) |
| 0.0 | 5.2 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 1.0 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 0.8 | GO:0070233 | negative regulation of T cell apoptotic process(GO:0070233) |
| 0.0 | 3.5 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 1.0 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.5 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.0 | 1.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 1.7 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.7 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.1 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.1 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 1.0 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.0 | 0.3 | GO:0035812 | renal sodium excretion(GO:0035812) regulation of renal sodium excretion(GO:0035813) |
| 0.0 | 0.9 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.1 | GO:0071748 | IgA immunoglobulin complex(GO:0071745) IgA immunoglobulin complex, circulating(GO:0071746) monomeric IgA immunoglobulin complex(GO:0071748) polymeric IgA immunoglobulin complex(GO:0071749) secretory IgA immunoglobulin complex(GO:0071751) |
| 1.1 | 10.7 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.3 | 6.0 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.3 | 5.6 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.2 | 2.0 | GO:0032039 | integrator complex(GO:0032039) |
| 0.2 | 9.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.2 | 3.6 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.9 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.9 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 2.8 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 1.7 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 1.4 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 1.0 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 1.1 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 5.3 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 1.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 1.0 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 4.8 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.0 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.4 | 1.2 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.3 | 1.0 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.2 | 9.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.2 | 1.3 | GO:0032396 | inhibitory MHC class I receptor activity(GO:0032396) |
| 0.1 | 1.4 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 0.4 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 0.7 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.1 | 2.8 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 5.6 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.1 | 2.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.4 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 1.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 10.0 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 1.1 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 0.3 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.8 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 5.2 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 1.1 | GO:0034061 | DNA-directed DNA polymerase activity(GO:0003887) DNA polymerase activity(GO:0034061) |
| 0.0 | 3.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.9 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.2 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 1.7 | GO:0005178 | integrin binding(GO:0005178) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 3.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.0 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.3 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.4 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.5 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 4.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 2.8 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 1.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.0 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.7 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 2.4 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.0 | 0.9 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 1.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |