Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
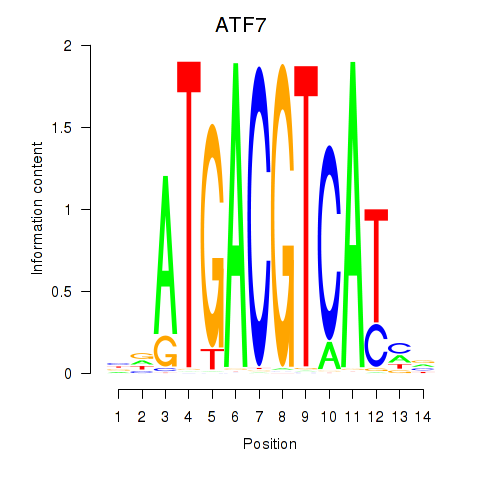
Results for ATF7
Z-value: 0.36
Transcription factors associated with ATF7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ATF7
|
ENSG00000170653.14 | activating transcription factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ATF7 | hg19_v2_chr12_-_54020107_54020199, hg19_v2_chr12_-_53994805_53994817 | 0.14 | 3.4e-02 | Click! |
Activity profile of ATF7 motif
Sorted Z-values of ATF7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_76829441 | 9.86 |
ENST00000458297.2
|
ATP9B
|
ATPase, class II, type 9B |
| chr1_+_26496362 | 7.92 |
ENST00000374266.5
ENST00000270812.5 |
ZNF593
|
zinc finger protein 593 |
| chr11_+_12695944 | 7.68 |
ENST00000361905.4
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr15_-_91537723 | 7.31 |
ENST00000394249.3
ENST00000559811.1 ENST00000442656.2 ENST00000557905.1 ENST00000361919.3 |
PRC1
|
protein regulator of cytokinesis 1 |
| chr11_+_12696102 | 6.86 |
ENST00000527636.1
ENST00000527376.1 |
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr4_-_122744998 | 6.09 |
ENST00000274026.5
|
CCNA2
|
cyclin A2 |
| chr19_+_10362577 | 6.08 |
ENST00000592514.1
ENST00000307422.5 ENST00000253099.6 ENST00000590150.1 ENST00000590669.1 |
MRPL4
|
mitochondrial ribosomal protein L4 |
| chr22_-_44258360 | 5.91 |
ENST00000330884.4
ENST00000249130.5 |
SULT4A1
|
sulfotransferase family 4A, member 1 |
| chr6_-_3157760 | 5.82 |
ENST00000333628.3
|
TUBB2A
|
tubulin, beta 2A class IIa |
| chr16_+_81040794 | 5.67 |
ENST00000439957.3
ENST00000393335.3 ENST00000428963.2 ENST00000564669.1 |
CENPN
|
centromere protein N |
| chr19_+_10362882 | 5.27 |
ENST00000393733.2
ENST00000588502.1 |
MRPL4
|
mitochondrial ribosomal protein L4 |
| chr16_-_3767506 | 5.26 |
ENST00000538171.1
|
TRAP1
|
TNF receptor-associated protein 1 |
| chr16_-_3767551 | 5.24 |
ENST00000246957.5
|
TRAP1
|
TNF receptor-associated protein 1 |
| chr8_+_98788003 | 5.22 |
ENST00000521545.2
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta |
| chr4_-_104119528 | 4.92 |
ENST00000380026.3
ENST00000503705.1 ENST00000265148.3 |
CENPE
|
centromere protein E, 312kDa |
| chr17_+_7155556 | 4.87 |
ENST00000570500.1
ENST00000574993.1 ENST00000396628.2 ENST00000573657.1 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr14_+_68086515 | 4.84 |
ENST00000261783.3
|
ARG2
|
arginase 2 |
| chr16_+_2039946 | 4.63 |
ENST00000248121.2
ENST00000568896.1 |
SYNGR3
|
synaptogyrin 3 |
| chr16_+_3068393 | 4.52 |
ENST00000573001.1
|
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A |
| chr7_+_5632436 | 4.40 |
ENST00000340250.6
ENST00000382361.3 |
FSCN1
|
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) |
| chr7_+_56019486 | 4.21 |
ENST00000446692.1
ENST00000285298.4 ENST00000443449.1 |
GBAS
MRPS17
|
glioblastoma amplified sequence mitochondrial ribosomal protein S17 |
| chr1_+_156030937 | 4.17 |
ENST00000361084.5
|
RAB25
|
RAB25, member RAS oncogene family |
| chr2_-_97760576 | 4.17 |
ENST00000414820.1
ENST00000272610.3 |
FAHD2B
|
fumarylacetoacetate hydrolase domain containing 2B |
| chr11_+_18343800 | 4.14 |
ENST00000453096.2
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa |
| chr14_+_93389425 | 4.11 |
ENST00000216492.5
ENST00000334654.4 |
CHGA
|
chromogranin A (parathyroid secretory protein 1) |
| chr22_+_32340481 | 4.05 |
ENST00000397492.1
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chr11_+_4116005 | 3.97 |
ENST00000300738.5
|
RRM1
|
ribonucleotide reductase M1 |
| chrX_+_110339439 | 3.96 |
ENST00000372010.1
ENST00000519681.1 ENST00000372007.5 |
PAK3
|
p21 protein (Cdc42/Rac)-activated kinase 3 |
| chr4_+_108910870 | 3.95 |
ENST00000403312.1
ENST00000603302.1 ENST00000309522.3 |
HADH
|
hydroxyacyl-CoA dehydrogenase |
| chr11_+_4116054 | 3.94 |
ENST00000423050.2
|
RRM1
|
ribonucleotide reductase M1 |
| chr16_+_29911864 | 3.83 |
ENST00000308748.5
|
ASPHD1
|
aspartate beta-hydroxylase domain containing 1 |
| chr4_+_108911036 | 3.81 |
ENST00000505878.1
|
HADH
|
hydroxyacyl-CoA dehydrogenase |
| chrX_-_47479246 | 3.78 |
ENST00000295987.7
ENST00000340666.4 |
SYN1
|
synapsin I |
| chr17_+_8152590 | 3.70 |
ENST00000584044.1
ENST00000314666.6 ENST00000545834.1 ENST00000581242.1 |
PFAS
|
phosphoribosylformylglycinamidine synthase |
| chr6_-_31774714 | 3.61 |
ENST00000375661.5
|
LSM2
|
LSM2 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr22_+_32340447 | 3.56 |
ENST00000248975.5
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chr20_+_42295745 | 3.47 |
ENST00000396863.4
ENST00000217026.4 |
MYBL2
|
v-myb avian myeloblastosis viral oncogene homolog-like 2 |
| chr1_-_16482554 | 3.34 |
ENST00000358432.5
|
EPHA2
|
EPH receptor A2 |
| chr17_+_7155343 | 3.32 |
ENST00000573513.1
ENST00000354429.2 ENST00000574255.1 ENST00000396627.2 ENST00000356683.2 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr19_+_57831829 | 3.26 |
ENST00000321545.4
|
ZNF543
|
zinc finger protein 543 |
| chrX_+_150148976 | 3.23 |
ENST00000419110.1
|
HMGB3
|
high mobility group box 3 |
| chr11_-_28129656 | 3.22 |
ENST00000263181.6
|
KIF18A
|
kinesin family member 18A |
| chr4_-_146019335 | 3.20 |
ENST00000451299.2
ENST00000507656.1 ENST00000309439.5 |
ANAPC10
|
anaphase promoting complex subunit 10 |
| chr18_+_2571510 | 3.18 |
ENST00000261597.4
ENST00000575515.1 |
NDC80
|
NDC80 kinetochore complex component |
| chr12_+_112856690 | 3.09 |
ENST00000392597.1
ENST00000351677.2 |
PTPN11
|
protein tyrosine phosphatase, non-receptor type 11 |
| chr4_+_85504075 | 3.08 |
ENST00000295887.5
|
CDS1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 |
| chr11_-_1593150 | 3.03 |
ENST00000397374.3
|
DUSP8
|
dual specificity phosphatase 8 |
| chr7_-_73153178 | 3.02 |
ENST00000437775.2
ENST00000222800.3 |
ABHD11
|
abhydrolase domain containing 11 |
| chr19_+_8455200 | 3.02 |
ENST00000601897.1
ENST00000594216.1 |
RAB11B
|
RAB11B, member RAS oncogene family |
| chr9_-_99382065 | 2.99 |
ENST00000265659.2
ENST00000375241.1 ENST00000375236.1 |
CDC14B
|
cell division cycle 14B |
| chr3_+_44803209 | 2.98 |
ENST00000326047.4
|
KIF15
|
kinesin family member 15 |
| chr16_+_56225248 | 2.96 |
ENST00000262493.6
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O |
| chr20_+_5892037 | 2.94 |
ENST00000378961.4
|
CHGB
|
chromogranin B (secretogranin 1) |
| chr17_-_7165662 | 2.90 |
ENST00000571881.2
ENST00000360325.7 |
CLDN7
|
claudin 7 |
| chr16_+_29911666 | 2.84 |
ENST00000563177.1
ENST00000483405.1 |
ASPHD1
|
aspartate beta-hydroxylase domain containing 1 |
| chr17_+_53342311 | 2.82 |
ENST00000226067.5
|
HLF
|
hepatic leukemia factor |
| chr19_+_8455077 | 2.77 |
ENST00000328024.6
|
RAB11B
|
RAB11B, member RAS oncogene family |
| chr11_-_33183006 | 2.73 |
ENST00000524827.1
ENST00000323959.4 ENST00000431742.2 |
CSTF3
|
cleavage stimulation factor, 3' pre-RNA, subunit 3, 77kDa |
| chr4_-_146019287 | 2.66 |
ENST00000502847.1
ENST00000513054.1 |
ANAPC10
|
anaphase promoting complex subunit 10 |
| chr14_+_88851874 | 2.64 |
ENST00000393545.4
ENST00000356583.5 ENST00000555401.1 ENST00000553885.1 |
SPATA7
|
spermatogenesis associated 7 |
| chr4_+_128802016 | 2.61 |
ENST00000270861.5
ENST00000515069.1 ENST00000513090.1 ENST00000507249.1 |
PLK4
|
polo-like kinase 4 |
| chr5_-_138210977 | 2.60 |
ENST00000274711.6
ENST00000521094.2 |
LRRTM2
|
leucine rich repeat transmembrane neuronal 2 |
| chr10_+_123922941 | 2.57 |
ENST00000360561.3
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr19_-_10341948 | 2.57 |
ENST00000590320.1
ENST00000592342.1 ENST00000588952.1 |
S1PR2
DNMT1
|
sphingosine-1-phosphate receptor 2 DNA (cytosine-5-)-methyltransferase 1 |
| chr11_+_18344106 | 2.53 |
ENST00000534641.1
ENST00000525831.1 ENST00000265963.4 |
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa |
| chr4_+_75310851 | 2.49 |
ENST00000395748.3
ENST00000264487.2 |
AREG
|
amphiregulin |
| chr11_+_3819049 | 2.46 |
ENST00000396986.2
ENST00000300730.6 ENST00000532535.1 ENST00000396993.4 ENST00000396991.2 ENST00000532523.1 ENST00000459679.1 ENST00000464261.1 ENST00000464906.2 ENST00000464441.1 |
PGAP2
|
post-GPI attachment to proteins 2 |
| chr10_+_123923105 | 2.45 |
ENST00000368999.1
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr19_-_460996 | 2.43 |
ENST00000264554.6
|
SHC2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr2_-_240964716 | 2.38 |
ENST00000404554.1
ENST00000407129.3 ENST00000307300.4 ENST00000443626.1 ENST00000252711.2 |
NDUFA10
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kDa |
| chr4_-_146019693 | 2.37 |
ENST00000514390.1
|
ANAPC10
|
anaphase promoting complex subunit 10 |
| chr17_+_33914460 | 2.36 |
ENST00000537622.2
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr4_+_75311019 | 2.34 |
ENST00000502307.1
|
AREG
|
amphiregulin |
| chr19_+_18723660 | 2.33 |
ENST00000262817.3
|
TMEM59L
|
transmembrane protein 59-like |
| chr19_+_13056663 | 2.32 |
ENST00000541222.1
ENST00000316856.3 ENST00000586534.1 ENST00000592268.1 |
RAD23A
|
RAD23 homolog A (S. cerevisiae) |
| chr15_-_66790146 | 2.29 |
ENST00000316634.5
|
SNAPC5
|
small nuclear RNA activating complex, polypeptide 5, 19kDa |
| chr2_+_149632783 | 2.28 |
ENST00000435030.1
|
KIF5C
|
kinesin family member 5C |
| chr16_-_81040719 | 2.26 |
ENST00000219400.3
|
CMC2
|
C-x(9)-C motif containing 2 |
| chr17_+_33914276 | 2.26 |
ENST00000592545.1
ENST00000538556.1 ENST00000312678.8 ENST00000589344.1 |
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr14_+_105219437 | 2.23 |
ENST00000329967.6
ENST00000347067.5 ENST00000553810.1 |
SIVA1
|
SIVA1, apoptosis-inducing factor |
| chr7_-_44924939 | 2.23 |
ENST00000395699.2
|
PURB
|
purine-rich element binding protein B |
| chr17_+_29421987 | 2.20 |
ENST00000431387.4
|
NF1
|
neurofibromin 1 |
| chr19_+_35849362 | 2.19 |
ENST00000327809.4
|
FFAR3
|
free fatty acid receptor 3 |
| chr17_+_29421900 | 2.16 |
ENST00000358273.4
ENST00000356175.3 |
NF1
|
neurofibromin 1 |
| chr4_-_668108 | 2.16 |
ENST00000304312.4
|
ATP5I
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit E |
| chr10_+_123923205 | 2.14 |
ENST00000369004.3
ENST00000260733.3 |
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr11_-_123525289 | 2.13 |
ENST00000392770.2
ENST00000299333.3 ENST00000530277.1 |
SCN3B
|
sodium channel, voltage-gated, type III, beta subunit |
| chr1_-_16345245 | 2.11 |
ENST00000311890.9
|
HSPB7
|
heat shock 27kDa protein family, member 7 (cardiovascular) |
| chr17_+_43209967 | 2.11 |
ENST00000431281.1
ENST00000591859.1 |
ACBD4
|
acyl-CoA binding domain containing 4 |
| chr19_+_45504688 | 2.10 |
ENST00000221452.8
ENST00000540120.1 ENST00000505236.1 |
RELB
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr7_-_140624499 | 2.07 |
ENST00000288602.6
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B |
| chr7_+_140396946 | 2.06 |
ENST00000476470.1
ENST00000471136.1 |
NDUFB2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
| chr19_+_52693259 | 2.02 |
ENST00000322088.6
ENST00000454220.2 ENST00000444322.2 ENST00000477989.1 |
PPP2R1A
|
protein phosphatase 2, regulatory subunit A, alpha |
| chr1_-_1284730 | 2.02 |
ENST00000378888.5
|
DVL1
|
dishevelled segment polarity protein 1 |
| chr9_-_131644202 | 1.98 |
ENST00000320665.6
ENST00000436267.2 |
CCBL1
|
cysteine conjugate-beta lyase, cytoplasmic |
| chr2_-_220174166 | 1.95 |
ENST00000409251.3
ENST00000451506.1 ENST00000295718.2 ENST00000446182.1 |
PTPRN
|
protein tyrosine phosphatase, receptor type, N |
| chr7_+_100464760 | 1.94 |
ENST00000200457.4
|
TRIP6
|
thyroid hormone receptor interactor 6 |
| chr3_+_170075436 | 1.93 |
ENST00000476188.1
ENST00000259119.4 ENST00000426052.2 |
SKIL
|
SKI-like oncogene |
| chr19_+_36103631 | 1.93 |
ENST00000203166.5
ENST00000379045.2 |
HAUS5
|
HAUS augmin-like complex, subunit 5 |
| chr1_+_70820451 | 1.91 |
ENST00000361764.4
ENST00000359875.5 ENST00000370940.5 ENST00000531950.1 ENST00000432224.1 |
HHLA3
|
HERV-H LTR-associating 3 |
| chr17_-_39780819 | 1.90 |
ENST00000311208.8
|
KRT17
|
keratin 17 |
| chr7_+_140396756 | 1.88 |
ENST00000460088.1
ENST00000472695.1 |
NDUFB2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
| chr3_+_158288999 | 1.84 |
ENST00000482628.1
ENST00000478894.2 ENST00000392822.3 ENST00000466246.1 |
MLF1
|
myeloid leukemia factor 1 |
| chr20_+_34203794 | 1.82 |
ENST00000374273.3
|
SPAG4
|
sperm associated antigen 4 |
| chr17_+_685513 | 1.79 |
ENST00000304478.4
|
RNMTL1
|
RNA methyltransferase like 1 |
| chr15_+_75287861 | 1.76 |
ENST00000425597.3
ENST00000562327.1 ENST00000568018.1 ENST00000562212.1 ENST00000567920.1 ENST00000566872.1 ENST00000361900.6 ENST00000545456.1 |
SCAMP5
|
secretory carrier membrane protein 5 |
| chr7_-_129592471 | 1.74 |
ENST00000473814.2
ENST00000490974.1 |
UBE2H
|
ubiquitin-conjugating enzyme E2H |
| chr5_+_131892603 | 1.73 |
ENST00000378823.3
ENST00000265335.6 |
RAD50
|
RAD50 homolog (S. cerevisiae) |
| chr19_+_36119929 | 1.73 |
ENST00000588161.1
ENST00000262633.4 ENST00000592202.1 ENST00000586618.1 |
RBM42
|
RNA binding motif protein 42 |
| chr1_-_70671216 | 1.72 |
ENST00000370952.3
|
LRRC40
|
leucine rich repeat containing 40 |
| chr19_+_50180317 | 1.70 |
ENST00000534465.1
|
PRMT1
|
protein arginine methyltransferase 1 |
| chr11_+_82783097 | 1.68 |
ENST00000501011.2
ENST00000527627.1 ENST00000526795.1 ENST00000533528.1 ENST00000533708.1 ENST00000534499.1 |
RAB30-AS1
|
RAB30 antisense RNA 1 (head to head) |
| chr17_+_68071458 | 1.68 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr17_+_41476327 | 1.68 |
ENST00000320033.4
|
ARL4D
|
ADP-ribosylation factor-like 4D |
| chr20_-_3154162 | 1.67 |
ENST00000360342.3
|
LZTS3
|
Homo sapiens leucine zipper, putative tumor suppressor family member 3 (LZTS3), mRNA. |
| chr2_-_224467093 | 1.67 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chr19_-_56826157 | 1.64 |
ENST00000592509.1
ENST00000592679.1 ENST00000588442.1 ENST00000593106.1 ENST00000587492.1 ENST00000254165.3 |
ZSCAN5A
|
zinc finger and SCAN domain containing 5A |
| chr19_+_36119975 | 1.64 |
ENST00000589559.1
ENST00000360475.4 |
RBM42
|
RNA binding motif protein 42 |
| chr12_-_58165870 | 1.59 |
ENST00000257848.7
|
METTL1
|
methyltransferase like 1 |
| chr22_+_25003626 | 1.58 |
ENST00000451366.1
ENST00000406383.2 ENST00000428855.1 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr4_+_75480629 | 1.58 |
ENST00000380846.3
|
AREGB
|
amphiregulin B |
| chr6_+_28048753 | 1.57 |
ENST00000377325.1
|
ZNF165
|
zinc finger protein 165 |
| chr4_-_2010562 | 1.57 |
ENST00000411649.1
ENST00000542778.1 ENST00000411638.2 ENST00000431323.1 |
NELFA
|
negative elongation factor complex member A |
| chr2_-_114514181 | 1.56 |
ENST00000409342.1
|
SLC35F5
|
solute carrier family 35, member F5 |
| chr19_-_8008533 | 1.56 |
ENST00000597926.1
|
TIMM44
|
translocase of inner mitochondrial membrane 44 homolog (yeast) |
| chr1_-_202896310 | 1.54 |
ENST00000367261.3
|
KLHL12
|
kelch-like family member 12 |
| chr11_-_31839488 | 1.54 |
ENST00000419022.1
ENST00000379132.3 ENST00000379129.2 |
PAX6
|
paired box 6 |
| chr11_+_66610883 | 1.52 |
ENST00000309657.3
ENST00000524506.1 |
RCE1
|
Ras converting CAAX endopeptidase 1 |
| chr12_-_93836028 | 1.51 |
ENST00000318066.2
|
UBE2N
|
ubiquitin-conjugating enzyme E2N |
| chr12_-_93835665 | 1.51 |
ENST00000552442.1
ENST00000550657.1 |
UBE2N
|
ubiquitin-conjugating enzyme E2N |
| chr9_-_2844058 | 1.51 |
ENST00000397885.2
|
KIAA0020
|
KIAA0020 |
| chr11_-_62389449 | 1.50 |
ENST00000534026.1
|
B3GAT3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr9_-_131644306 | 1.49 |
ENST00000302586.3
|
CCBL1
|
cysteine conjugate-beta lyase, cytoplasmic |
| chr9_-_33001520 | 1.48 |
ENST00000463596.1
ENST00000379819.1 ENST00000397172.3 ENST00000379812.5 ENST00000477119.1 ENST00000379813.3 ENST00000379825.2 ENST00000309615.3 ENST00000476858.1 ENST00000473221.1 |
APTX
|
aprataxin |
| chr17_+_68071389 | 1.47 |
ENST00000283936.1
ENST00000392671.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr19_+_36486078 | 1.47 |
ENST00000378887.2
|
SDHAF1
|
succinate dehydrogenase complex assembly factor 1 |
| chr1_-_115124257 | 1.46 |
ENST00000369541.3
|
BCAS2
|
breast carcinoma amplified sequence 2 |
| chr4_+_159131346 | 1.46 |
ENST00000508243.1
ENST00000296529.6 |
TMEM144
|
transmembrane protein 144 |
| chr12_-_56694142 | 1.46 |
ENST00000550655.1
ENST00000548567.1 ENST00000551430.2 ENST00000351328.3 |
CS
|
citrate synthase |
| chr14_+_96671016 | 1.43 |
ENST00000542454.2
ENST00000554311.1 ENST00000306005.3 ENST00000539359.1 ENST00000553811.1 |
BDKRB2
RP11-404P21.8
|
bradykinin receptor B2 Uncharacterized protein |
| chr14_+_93651296 | 1.43 |
ENST00000283534.4
ENST00000557574.1 |
TMEM251
RP11-371E8.4
|
transmembrane protein 251 Uncharacterized protein |
| chr14_+_93651358 | 1.41 |
ENST00000415050.2
|
TMEM251
|
transmembrane protein 251 |
| chr10_-_97416400 | 1.40 |
ENST00000371224.2
ENST00000371221.3 |
ALDH18A1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr3_+_158288942 | 1.38 |
ENST00000491767.1
ENST00000355893.5 |
MLF1
|
myeloid leukemia factor 1 |
| chr10_+_134351319 | 1.37 |
ENST00000368594.3
ENST00000368593.3 |
INPP5A
|
inositol polyphosphate-5-phosphatase, 40kDa |
| chr3_+_158288960 | 1.37 |
ENST00000484955.1
ENST00000359117.5 ENST00000498592.1 ENST00000477042.1 ENST00000471745.1 ENST00000469452.1 |
MLF1
|
myeloid leukemia factor 1 |
| chr19_+_44100632 | 1.37 |
ENST00000533118.1
|
ZNF576
|
zinc finger protein 576 |
| chr6_+_27833034 | 1.35 |
ENST00000357320.2
|
HIST1H2AL
|
histone cluster 1, H2al |
| chr1_+_28655505 | 1.35 |
ENST00000373842.4
ENST00000398997.2 |
MED18
|
mediator complex subunit 18 |
| chr8_-_82754427 | 1.34 |
ENST00000353788.4
ENST00000520618.1 ENST00000518183.1 ENST00000396330.2 ENST00000519119.1 ENST00000345957.4 |
SNX16
|
sorting nexin 16 |
| chr3_-_187388173 | 1.34 |
ENST00000287641.3
|
SST
|
somatostatin |
| chr3_-_107941230 | 1.33 |
ENST00000264538.3
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr3_+_62304712 | 1.33 |
ENST00000494481.1
|
C3orf14
|
chromosome 3 open reading frame 14 |
| chr9_-_111696340 | 1.29 |
ENST00000374647.5
|
IKBKAP
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase complex-associated protein |
| chr7_-_31380502 | 1.29 |
ENST00000297142.3
|
NEUROD6
|
neuronal differentiation 6 |
| chr1_-_23495340 | 1.29 |
ENST00000418342.1
|
LUZP1
|
leucine zipper protein 1 |
| chr21_+_45209394 | 1.28 |
ENST00000497547.1
|
RRP1
|
ribosomal RNA processing 1 |
| chr12_+_7014064 | 1.23 |
ENST00000443597.2
|
LRRC23
|
leucine rich repeat containing 23 |
| chr12_+_7013897 | 1.22 |
ENST00000007969.8
ENST00000323702.5 |
LRRC23
|
leucine rich repeat containing 23 |
| chr10_-_27444143 | 1.20 |
ENST00000477432.1
|
YME1L1
|
YME1-like 1 ATPase |
| chr11_+_108535752 | 1.20 |
ENST00000322536.3
|
DDX10
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 10 |
| chr4_-_186456652 | 1.18 |
ENST00000284767.5
ENST00000284770.5 |
PDLIM3
|
PDZ and LIM domain 3 |
| chr15_-_66790047 | 1.17 |
ENST00000566658.1
ENST00000563480.2 ENST00000395589.2 ENST00000307979.7 |
SNAPC5
|
small nuclear RNA activating complex, polypeptide 5, 19kDa |
| chr4_+_1873100 | 1.16 |
ENST00000508803.1
|
WHSC1
|
Wolf-Hirschhorn syndrome candidate 1 |
| chr1_+_110527308 | 1.15 |
ENST00000369799.5
|
AHCYL1
|
adenosylhomocysteinase-like 1 |
| chr2_+_3705785 | 1.15 |
ENST00000252505.3
|
ALLC
|
allantoicase |
| chr7_+_23221438 | 1.15 |
ENST00000258742.5
|
NUPL2
|
nucleoporin like 2 |
| chr9_-_99381660 | 1.15 |
ENST00000375240.3
ENST00000463569.1 |
CDC14B
|
cell division cycle 14B |
| chr11_-_13517565 | 1.14 |
ENST00000282091.1
ENST00000529816.1 |
PTH
|
parathyroid hormone |
| chr7_+_140396465 | 1.14 |
ENST00000476279.1
ENST00000247866.4 ENST00000461457.1 ENST00000465506.1 ENST00000204307.5 ENST00000464566.1 |
NDUFB2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
| chr9_+_97488939 | 1.13 |
ENST00000277198.2
ENST00000297979.5 |
C9orf3
|
chromosome 9 open reading frame 3 |
| chr3_+_62304648 | 1.12 |
ENST00000462069.1
ENST00000232519.5 ENST00000465142.1 |
C3orf14
|
chromosome 3 open reading frame 14 |
| chrX_-_7895755 | 1.11 |
ENST00000444736.1
ENST00000537427.1 ENST00000442940.1 |
PNPLA4
|
patatin-like phospholipase domain containing 4 |
| chr19_+_36036583 | 1.10 |
ENST00000392205.1
|
TMEM147
|
transmembrane protein 147 |
| chr11_-_68039364 | 1.09 |
ENST00000533310.1
ENST00000304271.6 ENST00000527280.1 |
C11orf24
|
chromosome 11 open reading frame 24 |
| chr16_+_19079311 | 1.09 |
ENST00000569127.1
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr4_-_79860506 | 1.08 |
ENST00000295462.3
ENST00000380645.4 ENST00000512733.1 |
PAQR3
|
progestin and adipoQ receptor family member III |
| chr10_+_35484793 | 1.07 |
ENST00000488741.1
ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chr3_-_48130707 | 1.06 |
ENST00000360240.6
ENST00000383737.4 |
MAP4
|
microtubule-associated protein 4 |
| chr16_-_3068171 | 1.05 |
ENST00000572154.1
ENST00000328796.4 |
CLDN6
|
claudin 6 |
| chr17_+_41177220 | 1.05 |
ENST00000587250.2
ENST00000544533.1 |
RND2
|
Rho family GTPase 2 |
| chr4_-_186456766 | 1.04 |
ENST00000284771.6
|
PDLIM3
|
PDZ and LIM domain 3 |
| chr12_+_132628963 | 1.03 |
ENST00000330579.1
|
NOC4L
|
nucleolar complex associated 4 homolog (S. cerevisiae) |
| chr19_+_36120009 | 1.01 |
ENST00000589871.1
|
RBM42
|
RNA binding motif protein 42 |
| chr2_-_220408430 | 1.00 |
ENST00000243776.6
|
CHPF
|
chondroitin polymerizing factor |
| chr4_+_668348 | 1.00 |
ENST00000511290.1
|
MYL5
|
myosin, light chain 5, regulatory |
| chr18_+_23806437 | 0.99 |
ENST00000578121.1
|
TAF4B
|
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa |
| chr1_+_17531614 | 0.99 |
ENST00000375471.4
|
PADI1
|
peptidyl arginine deiminase, type I |
| chr9_-_111696224 | 0.98 |
ENST00000537196.1
|
IKBKAP
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase complex-associated protein |
| chr3_-_119182523 | 0.97 |
ENST00000319172.5
|
TMEM39A
|
transmembrane protein 39A |
| chr19_+_57874835 | 0.95 |
ENST00000543226.1
ENST00000596755.1 ENST00000282282.3 ENST00000597658.1 |
TRAPPC2P1
ZNF547
AC003002.4
|
trafficking protein particle complex 2 pseudogene 1 zinc finger protein 547 Uncharacterized protein |
| chr22_+_31518938 | 0.95 |
ENST00000412985.1
ENST00000331075.5 ENST00000412277.2 ENST00000420017.1 ENST00000400294.2 ENST00000405300.1 ENST00000404390.3 |
INPP5J
|
inositol polyphosphate-5-phosphatase J |
| chr1_+_2005425 | 0.94 |
ENST00000461106.2
|
PRKCZ
|
protein kinase C, zeta |
| chr7_-_129592700 | 0.94 |
ENST00000472396.1
ENST00000355621.3 |
UBE2H
|
ubiquitin-conjugating enzyme E2H |
| chr12_-_121454148 | 0.94 |
ENST00000535367.1
ENST00000538296.1 ENST00000445832.3 ENST00000536407.2 ENST00000366211.2 ENST00000539736.1 ENST00000288757.3 ENST00000537817.1 |
C12orf43
|
chromosome 12 open reading frame 43 |
| chr6_-_26285737 | 0.93 |
ENST00000377727.1
ENST00000289352.1 |
HIST1H4H
|
histone cluster 1, H4h |
| chr2_-_21022818 | 0.93 |
ENST00000440866.2
ENST00000541941.1 ENST00000402479.2 ENST00000435420.2 ENST00000432947.1 ENST00000403006.2 ENST00000419825.2 ENST00000381090.3 ENST00000237822.3 ENST00000412261.1 |
C2orf43
|
chromosome 2 open reading frame 43 |
| chr1_+_6684918 | 0.92 |
ENST00000054650.4
|
THAP3
|
THAP domain containing, apoptosis associated protein 3 |
| chr19_+_49122548 | 0.92 |
ENST00000245222.4
ENST00000340932.3 ENST00000601712.1 ENST00000600537.1 |
SPHK2
|
sphingosine kinase 2 |
| chr7_-_124569991 | 0.92 |
ENST00000446993.1
ENST00000357628.3 ENST00000393329.1 |
POT1
|
protection of telomeres 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ATF7
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 10.5 | GO:0009386 | translational attenuation(GO:0009386) |
| 1.5 | 6.1 | GO:0071373 | cellular response to luteinizing hormone stimulus(GO:0071373) |
| 1.5 | 4.4 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 1.4 | 4.1 | GO:1901899 | dense core granule biogenesis(GO:0061110) positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of dense core granule biogenesis(GO:2000705) |
| 1.1 | 3.3 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 1.0 | 7.9 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.9 | 4.6 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.8 | 4.9 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.8 | 3.1 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.7 | 6.7 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.7 | 4.4 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.7 | 2.8 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.7 | 2.0 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.6 | 1.9 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.6 | 2.6 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.6 | 13.5 | GO:0051231 | spindle elongation(GO:0051231) |
| 0.6 | 4.8 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.6 | 5.8 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.6 | 1.7 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.5 | 2.6 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.5 | 1.6 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.5 | 7.6 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.4 | 3.1 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 0.4 | 4.8 | GO:0000050 | urea cycle(GO:0000050) |
| 0.4 | 1.7 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.4 | 3.8 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.4 | 1.9 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.4 | 1.9 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.4 | 1.2 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 0.4 | 1.1 | GO:1900158 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.4 | 2.3 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.4 | 3.7 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.4 | 3.0 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.4 | 3.2 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.3 | 7.4 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.3 | 12.8 | GO:1902895 | positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.3 | 6.7 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.3 | 1.5 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.3 | 2.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.3 | 2.4 | GO:0019240 | citrulline biosynthetic process(GO:0019240) |
| 0.3 | 2.6 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.3 | 4.2 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.3 | 1.6 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.3 | 7.1 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.3 | 2.3 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.2 | 10.9 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.2 | 1.4 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.2 | 3.0 | GO:0033182 | regulation of histone ubiquitination(GO:0033182) |
| 0.2 | 3.2 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.2 | 1.6 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.2 | 0.7 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.2 | 2.6 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.2 | 0.6 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 0.2 | 0.4 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.2 | 15.6 | GO:0070126 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.2 | 0.6 | GO:1903751 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.2 | 7.8 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.2 | 1.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.2 | 0.2 | GO:0009182 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) deoxyribonucleoside diphosphate metabolic process(GO:0009186) dGDP metabolic process(GO:0046066) |
| 0.2 | 5.7 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.2 | 1.4 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.2 | 1.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.2 | 1.5 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.2 | 5.9 | GO:0050427 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.2 | 1.9 | GO:0042635 | positive regulation of hair cycle(GO:0042635) positive regulation of hair follicle development(GO:0051798) |
| 0.2 | 3.0 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.2 | 0.3 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.1 | 4.0 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 4.0 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.1 | 2.0 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.1 | 0.4 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.5 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.1 | 1.8 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.1 | 0.2 | GO:0046878 | regulation of saliva secretion(GO:0046877) positive regulation of saliva secretion(GO:0046878) |
| 0.1 | 1.4 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 7.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.1 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.1 | 1.7 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.1 | 1.3 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.1 | 1.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 1.6 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 3.6 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 | 0.6 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.1 | 0.9 | GO:0051096 | telomere assembly(GO:0032202) positive regulation of helicase activity(GO:0051096) |
| 0.1 | 1.8 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.1 | 2.2 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 2.7 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.1 | 3.6 | GO:0043278 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.1 | 0.5 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.1 | 0.5 | GO:0034650 | aldosterone metabolic process(GO:0032341) aldosterone biosynthetic process(GO:0032342) cortisol metabolic process(GO:0034650) cortisol biosynthetic process(GO:0034651) |
| 0.1 | 0.7 | GO:0090520 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.1 | 7.2 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.1 | 3.4 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.1 | 4.6 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.1 | 2.8 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.1 | 4.5 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.1 | 2.3 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 3.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 2.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 1.3 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.1 | 0.6 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.1 | 1.3 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.1 | 0.9 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 3.5 | GO:0043525 | positive regulation of neuron apoptotic process(GO:0043525) |
| 0.0 | 1.9 | GO:0033108 | mitochondrial respiratory chain complex assembly(GO:0033108) |
| 0.0 | 1.5 | GO:0007507 | heart development(GO:0007507) |
| 0.0 | 0.3 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 1.3 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 1.4 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 1.3 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 1.5 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 1.5 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.3 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.4 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 1.1 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.7 | GO:0040020 | regulation of meiotic nuclear division(GO:0040020) |
| 0.0 | 0.2 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 1.8 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.9 | GO:0032201 | telomere maintenance via semi-conservative replication(GO:0032201) |
| 0.0 | 1.0 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 1.0 | GO:0051031 | tRNA export from nucleus(GO:0006409) tRNA transport(GO:0051031) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 1.1 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 1.4 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.4 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 2.4 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.0 | 1.0 | GO:0048477 | oogenesis(GO:0048477) |
| 0.0 | 0.5 | GO:0045666 | positive regulation of neuron differentiation(GO:0045666) |
| 0.0 | 2.2 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.0 | 0.9 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 3.2 | GO:0006310 | DNA recombination(GO:0006310) |
| 0.0 | 1.3 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.6 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.5 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.0 | 0.4 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 1.4 | GO:0007219 | Notch signaling pathway(GO:0007219) |
| 0.0 | 2.1 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.0 | 1.9 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.0 | 2.2 | GO:0045637 | regulation of myeloid cell differentiation(GO:0045637) |
| 0.0 | 0.3 | GO:0016578 | histone deubiquitination(GO:0016578) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 14.5 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 2.0 | 6.1 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 2.0 | 7.9 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 1.5 | 4.4 | GO:0044393 | microspike(GO:0044393) |
| 1.2 | 10.5 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 1.2 | 3.5 | GO:0031523 | Myb complex(GO:0031523) |
| 1.1 | 3.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 1.0 | 3.0 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 1.0 | 3.0 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.9 | 2.6 | GO:0098536 | deuterosome(GO:0098536) |
| 0.7 | 7.3 | GO:0070938 | contractile ring(GO:0070938) |
| 0.7 | 8.1 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.7 | 6.7 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.5 | 3.6 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.4 | 1.9 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.3 | 1.0 | GO:0030689 | Noc complex(GO:0030689) |
| 0.3 | 1.6 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.3 | 1.6 | GO:0032021 | NELF complex(GO:0032021) |
| 0.3 | 9.2 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.3 | 11.4 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.3 | 8.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.3 | 1.4 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.3 | 1.6 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.2 | 1.7 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.2 | 3.7 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.2 | 4.6 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.2 | 4.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 4.1 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.2 | 1.4 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 2.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.2 | 4.2 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.2 | 2.0 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 0.8 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 11.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 2.3 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 2.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.9 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 2.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 4.0 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.9 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 3.8 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 1.0 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 7.5 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 1.3 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.1 | 0.6 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.1 | 7.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 1.7 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 7.4 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 2.6 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 3.3 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 2.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.2 | GO:0020003 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.1 | 6.2 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 4.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 3.1 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 1.9 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 3.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0033011 | perinuclear theca(GO:0033011) cytoskeletal calyx(GO:0033150) |
| 0.0 | 2.3 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.1 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 3.4 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 1.1 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.3 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.4 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 15.6 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.3 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 3.5 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 1.7 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 2.6 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.6 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.3 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.3 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 2.3 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.9 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.4 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 4.3 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 2.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.9 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 1.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 2.8 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 4.0 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.4 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 6.1 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 0.5 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.2 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 1.0 | GO:0005643 | nuclear pore(GO:0005643) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.9 | GO:0000035 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 2.0 | 7.9 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 1.2 | 4.9 | GO:0043515 | kinetochore binding(GO:0043515) |
| 1.0 | 3.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.9 | 18.0 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.6 | 1.9 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.6 | 1.8 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.5 | 7.0 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.4 | 2.6 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.4 | 2.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.4 | 7.8 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.4 | 10.0 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.4 | 2.3 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.4 | 1.9 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.3 | 4.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.3 | 1.0 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.3 | 7.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.3 | 1.5 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.3 | 7.1 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.3 | 6.7 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.3 | 1.7 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.3 | 1.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.3 | 12.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.3 | 3.7 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.3 | 1.6 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.3 | 5.9 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.2 | 3.0 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.2 | 1.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.2 | 2.3 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.2 | 2.3 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.2 | 0.7 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.2 | 1.7 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.2 | 0.6 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.2 | 4.5 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.2 | 3.6 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.2 | 4.6 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.2 | 1.2 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.2 | 2.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 1.5 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.2 | 5.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 0.5 | GO:0047783 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 0.2 | 3.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 1.7 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.7 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 4.0 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 3.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.1 | 7.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.6 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.1 | 2.3 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 7.0 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 0.9 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 6.1 | GO:0097472 | cyclin-dependent protein kinase activity(GO:0097472) |
| 0.1 | 2.9 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 1.6 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 2.1 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 1.9 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 1.8 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.6 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 4.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.9 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.1 | 2.1 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 4.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 2.1 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 0.2 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.1 | 1.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 1.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 3.5 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.1 | 6.7 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.1 | 1.0 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 0.3 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 2.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 4.3 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 2.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.4 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 0.6 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 1.7 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.6 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.5 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.6 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 1.1 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 1.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 2.4 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 6.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 6.0 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 1.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 6.9 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 2.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 4.4 | GO:0008144 | drug binding(GO:0008144) |
| 0.0 | 1.5 | GO:1901476 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.0 | 2.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 3.0 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.0 | 2.1 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 2.5 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 2.5 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 2.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 3.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 1.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.2 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 1.5 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.3 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 5.1 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 4.6 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 2.7 | GO:0003924 | GTPase activity(GO:0003924) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 21.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.2 | 7.2 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.2 | 6.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 9.0 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 5.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 12.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 8.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 3.8 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.2 | 5.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 3.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 11.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 3.6 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 2.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 2.4 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 1.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 2.7 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 2.1 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 3.5 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 2.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.9 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.9 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 1.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.8 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.4 | 14.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.3 | 9.6 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.3 | 8.2 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.3 | 3.0 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.2 | 3.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.2 | 4.4 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.2 | 3.6 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.2 | 6.7 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.2 | 3.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 7.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 5.9 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 4.6 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.2 | 5.8 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.2 | 4.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 2.4 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.2 | 4.1 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.2 | 3.0 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.2 | 2.7 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 7.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 6.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 4.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 2.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 1.7 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 3.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 3.2 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.1 | 7.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 1.0 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 0.9 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 1.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 2.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.8 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 1.7 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 3.2 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.5 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 3.1 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 5.8 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.7 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.5 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 2.8 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 4.2 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 1.4 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |