Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
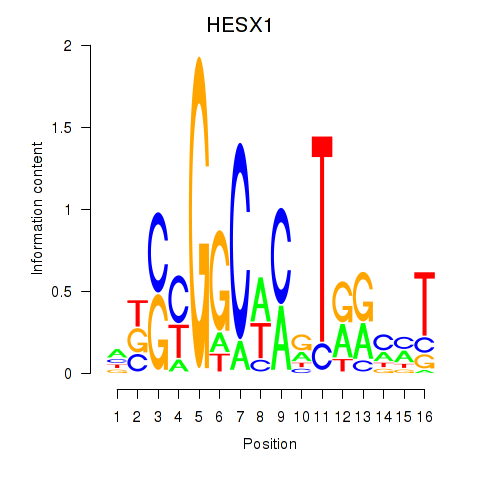
Results for HESX1
Z-value: 0.93
Transcription factors associated with HESX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HESX1
|
ENSG00000163666.4 | HESX homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HESX1 | hg19_v2_chr3_-_57233966_57234014 | -0.07 | 3.3e-01 | Click! |
Activity profile of HESX1 motif
Sorted Z-values of HESX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_106406090 | 31.91 |
ENST00000390593.2
|
IGHV6-1
|
immunoglobulin heavy variable 6-1 |
| chr2_-_89292422 | 31.72 |
ENST00000495489.1
|
IGKV1-8
|
immunoglobulin kappa variable 1-8 |
| chr2_+_90198535 | 31.58 |
ENST00000390276.2
|
IGKV1D-12
|
immunoglobulin kappa variable 1D-12 |
| chr2_+_90248739 | 26.58 |
ENST00000468879.1
|
IGKV1D-43
|
immunoglobulin kappa variable 1D-43 |
| chr2_-_89310012 | 25.51 |
ENST00000493819.1
|
IGKV1-9
|
immunoglobulin kappa variable 1-9 |
| chr2_-_89399845 | 24.25 |
ENST00000479981.1
|
IGKV1-16
|
immunoglobulin kappa variable 1-16 |
| chr2_-_89513402 | 23.59 |
ENST00000498435.1
|
IGKV1-27
|
immunoglobulin kappa variable 1-27 |
| chr2_-_89340242 | 23.39 |
ENST00000480492.1
|
IGKV1-12
|
immunoglobulin kappa variable 1-12 |
| chr2_+_89952792 | 14.44 |
ENST00000390265.2
|
IGKV1D-33
|
immunoglobulin kappa variable 1D-33 |
| chr2_+_90192768 | 13.29 |
ENST00000390275.2
|
IGKV1D-13
|
immunoglobulin kappa variable 1D-13 |
| chr2_-_89568263 | 12.36 |
ENST00000473726.1
|
IGKV1-33
|
immunoglobulin kappa variable 1-33 |
| chr2_+_114163945 | 10.71 |
ENST00000453673.3
|
IGKV1OR2-108
|
immunoglobulin kappa variable 1/OR2-108 (non-functional) |
| chr2_-_89247338 | 9.83 |
ENST00000496168.1
|
IGKV1-5
|
immunoglobulin kappa variable 1-5 |
| chr2_+_90458201 | 9.65 |
ENST00000603238.1
|
CH17-132F21.1
|
Uncharacterized protein |
| chr2_+_90259593 | 6.67 |
ENST00000471857.1
|
IGKV1D-8
|
immunoglobulin kappa variable 1D-8 |
| chr8_-_98290087 | 6.45 |
ENST00000322128.3
|
TSPYL5
|
TSPY-like 5 |
| chr4_-_2264015 | 5.89 |
ENST00000337190.2
|
MXD4
|
MAX dimerization protein 4 |
| chr1_+_159175201 | 5.05 |
ENST00000368121.2
|
DARC
|
Duffy blood group, atypical chemokine receptor |
| chr7_-_37024665 | 4.91 |
ENST00000396040.2
|
ELMO1
|
engulfment and cell motility 1 |
| chr14_-_25103472 | 4.69 |
ENST00000216341.4
ENST00000382542.1 ENST00000382540.1 |
GZMB
|
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr18_+_54318616 | 4.62 |
ENST00000254442.3
|
WDR7
|
WD repeat domain 7 |
| chr11_-_5276008 | 4.28 |
ENST00000336906.4
|
HBG2
|
hemoglobin, gamma G |
| chr18_+_54318566 | 4.16 |
ENST00000589935.1
ENST00000357574.3 |
WDR7
|
WD repeat domain 7 |
| chr10_-_101380121 | 4.12 |
ENST00000370495.4
|
SLC25A28
|
solute carrier family 25 (mitochondrial iron transporter), member 28 |
| chr14_-_25103388 | 3.92 |
ENST00000526004.1
ENST00000415355.3 |
GZMB
|
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr4_+_156588350 | 3.82 |
ENST00000296518.7
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr8_+_120885949 | 3.75 |
ENST00000523492.1
ENST00000286234.5 |
DEPTOR
|
DEP domain containing MTOR-interacting protein |
| chr1_-_203320617 | 3.75 |
ENST00000354955.4
|
FMOD
|
fibromodulin |
| chr20_-_590944 | 3.67 |
ENST00000246080.3
|
TCF15
|
transcription factor 15 (basic helix-loop-helix) |
| chr11_-_126138808 | 3.46 |
ENST00000332118.6
ENST00000532259.1 |
SRPR
|
signal recognition particle receptor (docking protein) |
| chr1_-_17216143 | 3.40 |
ENST00000457075.1
|
RP11-108M9.4
|
RP11-108M9.4 |
| chr2_-_43823119 | 3.34 |
ENST00000403856.1
ENST00000404790.1 ENST00000405975.2 ENST00000415080.2 |
THADA
|
thyroid adenoma associated |
| chr2_-_160654745 | 3.27 |
ENST00000259053.4
ENST00000429078.2 |
CD302
|
CD302 molecule |
| chr8_-_110656995 | 3.21 |
ENST00000276646.9
ENST00000533065.1 |
SYBU
|
syntabulin (syntaxin-interacting) |
| chr6_-_29600832 | 3.10 |
ENST00000377016.4
ENST00000376977.3 ENST00000377034.4 |
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr2_-_43823093 | 3.06 |
ENST00000405006.4
|
THADA
|
thyroid adenoma associated |
| chr16_+_4784273 | 3.01 |
ENST00000299320.5
ENST00000586724.1 |
C16orf71
|
chromosome 16 open reading frame 71 |
| chr14_+_61654271 | 2.96 |
ENST00000555185.1
ENST00000557294.1 ENST00000556778.1 |
PRKCH
|
protein kinase C, eta |
| chr4_+_71063641 | 2.95 |
ENST00000514097.1
|
ODAM
|
odontogenic, ameloblast asssociated |
| chr19_+_42388437 | 2.92 |
ENST00000378152.4
ENST00000337665.4 |
ARHGEF1
|
Rho guanine nucleotide exchange factor (GEF) 1 |
| chr19_+_19431490 | 2.91 |
ENST00000392313.6
ENST00000262815.8 ENST00000609122.1 |
MAU2
|
MAU2 sister chromatid cohesion factor |
| chr16_+_4784458 | 2.86 |
ENST00000590191.1
|
C16orf71
|
chromosome 16 open reading frame 71 |
| chr11_-_71791435 | 2.85 |
ENST00000351960.6
ENST00000541719.1 ENST00000535111.1 |
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr5_+_40679584 | 2.79 |
ENST00000302472.3
|
PTGER4
|
prostaglandin E receptor 4 (subtype EP4) |
| chr11_-_71791518 | 2.72 |
ENST00000537217.1
ENST00000366394.3 ENST00000358965.6 ENST00000546131.1 ENST00000543937.1 ENST00000368959.5 ENST00000541641.1 |
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr2_-_197036289 | 2.72 |
ENST00000263955.4
|
STK17B
|
serine/threonine kinase 17b |
| chr1_+_47799446 | 2.61 |
ENST00000371873.5
|
CMPK1
|
cytidine monophosphate (UMP-CMP) kinase 1, cytosolic |
| chr6_-_41888843 | 2.61 |
ENST00000434077.1
ENST00000409312.1 |
MED20
|
mediator complex subunit 20 |
| chr4_+_144303093 | 2.58 |
ENST00000505913.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr7_+_72742178 | 2.52 |
ENST00000442793.1
ENST00000413573.2 ENST00000252037.4 |
FKBP6
|
FK506 binding protein 6, 36kDa |
| chr11_+_125774258 | 2.48 |
ENST00000263576.6
|
DDX25
|
DEAD (Asp-Glu-Ala-Asp) box helicase 25 |
| chr1_-_40237020 | 2.45 |
ENST00000327582.5
|
OXCT2
|
3-oxoacid CoA transferase 2 |
| chr4_+_6784401 | 2.37 |
ENST00000425103.1
ENST00000307659.5 |
KIAA0232
|
KIAA0232 |
| chr16_-_57219966 | 2.28 |
ENST00000565760.1
ENST00000309137.8 ENST00000570184.1 ENST00000562324.1 |
FAM192A
|
family with sequence similarity 192, member A |
| chr18_-_54318353 | 2.25 |
ENST00000590954.1
ENST00000540155.1 |
TXNL1
|
thioredoxin-like 1 |
| chr7_-_95025661 | 2.25 |
ENST00000542556.1
ENST00000265627.5 ENST00000427422.1 ENST00000451904.1 |
PON1
PON3
|
paraoxonase 1 paraoxonase 3 |
| chr5_-_176057518 | 2.23 |
ENST00000393693.2
|
SNCB
|
synuclein, beta |
| chr13_-_52378231 | 2.17 |
ENST00000280056.2
ENST00000444610.2 |
DHRS12
|
dehydrogenase/reductase (SDR family) member 12 |
| chr16_-_1661988 | 2.09 |
ENST00000426508.2
|
IFT140
|
intraflagellar transport 140 homolog (Chlamydomonas) |
| chr14_-_90798418 | 2.08 |
ENST00000354366.3
|
NRDE2
|
NRDE-2, necessary for RNA interference, domain containing |
| chr8_+_27182862 | 2.05 |
ENST00000521164.1
ENST00000346049.5 |
PTK2B
|
protein tyrosine kinase 2 beta |
| chr22_+_38453378 | 1.97 |
ENST00000437453.1
ENST00000356976.3 |
PICK1
|
protein interacting with PRKCA 1 |
| chr16_-_70719925 | 1.97 |
ENST00000338779.6
|
MTSS1L
|
metastasis suppressor 1-like |
| chr16_+_1662326 | 1.97 |
ENST00000397412.3
|
CRAMP1L
|
Crm, cramped-like (Drosophila) |
| chr7_+_72742162 | 1.97 |
ENST00000431982.2
|
FKBP6
|
FK506 binding protein 6, 36kDa |
| chr1_-_20446020 | 1.88 |
ENST00000375105.3
|
PLA2G2D
|
phospholipase A2, group IID |
| chr11_+_73019282 | 1.82 |
ENST00000263674.3
|
ARHGEF17
|
Rho guanine nucleotide exchange factor (GEF) 17 |
| chr6_-_167275991 | 1.81 |
ENST00000510118.1
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr12_+_13197218 | 1.80 |
ENST00000197268.8
|
KIAA1467
|
KIAA1467 |
| chr6_+_31939608 | 1.78 |
ENST00000375331.2
ENST00000375333.2 |
STK19
|
serine/threonine kinase 19 |
| chr22_+_31892373 | 1.75 |
ENST00000443011.1
ENST00000400289.1 ENST00000444859.1 ENST00000400288.2 |
SFI1
|
Sfi1 homolog, spindle assembly associated (yeast) |
| chr16_+_66914264 | 1.73 |
ENST00000311765.2
ENST00000568869.1 ENST00000561704.1 ENST00000568398.1 ENST00000566776.1 |
PDP2
|
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
| chr2_-_128051670 | 1.73 |
ENST00000493187.2
|
ERCC3
|
excision repair cross-complementing rodent repair deficiency, complementation group 3 |
| chr6_-_46703069 | 1.70 |
ENST00000538237.1
ENST00000274793.7 |
PLA2G7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr12_-_752786 | 1.69 |
ENST00000433832.2
ENST00000542920.1 |
NINJ2
|
ninjurin 2 |
| chr21_+_38445539 | 1.56 |
ENST00000418766.1
ENST00000450533.1 ENST00000438055.1 ENST00000355666.1 ENST00000540756.1 ENST00000399010.1 |
TTC3
|
tetratricopeptide repeat domain 3 |
| chr3_+_178253993 | 1.54 |
ENST00000420517.2
ENST00000452583.1 |
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr17_+_45286387 | 1.40 |
ENST00000572316.1
ENST00000354968.1 ENST00000576874.1 ENST00000536623.2 |
MYL4
|
myosin, light chain 4, alkali; atrial, embryonic |
| chr6_-_41888814 | 1.40 |
ENST00000409060.1
ENST00000265350.4 |
MED20
|
mediator complex subunit 20 |
| chr11_-_57148619 | 1.39 |
ENST00000287143.2
|
PRG3
|
proteoglycan 3 |
| chr22_+_38453207 | 1.34 |
ENST00000404072.3
ENST00000424694.1 |
PICK1
|
protein interacting with PRKCA 1 |
| chr8_+_79428539 | 1.33 |
ENST00000352966.5
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha |
| chr7_+_74508372 | 1.30 |
ENST00000356115.5
ENST00000430511.2 ENST00000312575.7 |
GTF2IRD2B
|
GTF2I repeat domain containing 2B |
| chr17_+_18128896 | 1.30 |
ENST00000316843.4
|
LLGL1
|
lethal giant larvae homolog 1 (Drosophila) |
| chr10_-_75634326 | 1.28 |
ENST00000322635.3
ENST00000444854.2 ENST00000423381.1 ENST00000322680.3 ENST00000394762.2 |
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr8_+_97274119 | 1.27 |
ENST00000455950.2
|
PTDSS1
|
phosphatidylserine synthase 1 |
| chr3_+_158519654 | 1.27 |
ENST00000415822.2
ENST00000392813.4 ENST00000264266.8 |
MFSD1
|
major facilitator superfamily domain containing 1 |
| chr19_+_38397839 | 1.26 |
ENST00000222345.6
|
SIPA1L3
|
signal-induced proliferation-associated 1 like 3 |
| chr16_+_84733575 | 1.24 |
ENST00000219473.7
ENST00000563892.1 ENST00000562283.1 ENST00000570191.1 ENST00000569038.1 ENST00000570053.1 |
USP10
|
ubiquitin specific peptidase 10 |
| chr1_-_205290865 | 1.23 |
ENST00000367157.3
|
NUAK2
|
NUAK family, SNF1-like kinase, 2 |
| chr19_+_45312347 | 1.17 |
ENST00000270233.6
ENST00000591520.1 |
BCAM
|
basal cell adhesion molecule (Lutheran blood group) |
| chr21_-_38445443 | 1.17 |
ENST00000360525.4
|
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr2_-_96931679 | 1.13 |
ENST00000258439.3
ENST00000432959.1 |
TMEM127
|
transmembrane protein 127 |
| chr10_+_26505594 | 1.13 |
ENST00000259271.3
|
GAD2
|
glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa) |
| chr7_+_4815238 | 1.09 |
ENST00000348624.4
ENST00000401897.1 |
AP5Z1
|
adaptor-related protein complex 5, zeta 1 subunit |
| chrX_-_7895479 | 1.04 |
ENST00000381042.4
|
PNPLA4
|
patatin-like phospholipase domain containing 4 |
| chr2_+_176957619 | 0.99 |
ENST00000392539.3
|
HOXD13
|
homeobox D13 |
| chr14_-_91884150 | 0.99 |
ENST00000553403.1
|
CCDC88C
|
coiled-coil domain containing 88C |
| chr4_-_48782259 | 0.98 |
ENST00000507711.1
ENST00000358350.4 ENST00000537810.1 ENST00000264319.7 |
FRYL
|
FRY-like |
| chr21_+_34398153 | 0.95 |
ENST00000382357.3
ENST00000430860.1 ENST00000333337.3 |
OLIG2
|
oligodendrocyte lineage transcription factor 2 |
| chr8_-_101734308 | 0.94 |
ENST00000519004.1
ENST00000519363.1 ENST00000520142.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr7_-_74267836 | 0.85 |
ENST00000361071.5
ENST00000453619.2 ENST00000417115.2 ENST00000405086.2 |
GTF2IRD2
|
GTF2I repeat domain containing 2 |
| chr2_+_233562015 | 0.85 |
ENST00000427233.1
ENST00000373566.3 ENST00000373563.4 ENST00000428883.1 ENST00000456491.1 ENST00000409480.1 ENST00000421433.1 ENST00000425040.1 ENST00000430720.1 ENST00000409547.1 ENST00000423659.1 ENST00000409196.3 ENST00000409451.3 ENST00000429187.1 ENST00000440945.1 |
GIGYF2
|
GRB10 interacting GYF protein 2 |
| chr20_-_62587735 | 0.85 |
ENST00000354216.6
ENST00000369892.3 ENST00000358711.3 |
UCKL1
|
uridine-cytidine kinase 1-like 1 |
| chr17_+_45286706 | 0.83 |
ENST00000393450.1
ENST00000572303.1 |
MYL4
|
myosin, light chain 4, alkali; atrial, embryonic |
| chr11_+_64323156 | 0.80 |
ENST00000377585.3
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr20_+_18269121 | 0.78 |
ENST00000377671.3
ENST00000360010.5 ENST00000396026.3 ENST00000402618.2 ENST00000401790.1 ENST00000434018.1 ENST00000538547.1 ENST00000535822.1 |
ZNF133
|
zinc finger protein 133 |
| chr1_+_46016703 | 0.74 |
ENST00000481885.1
ENST00000351829.4 ENST00000471651.1 |
AKR1A1
|
aldo-keto reductase family 1, member A1 (aldehyde reductase) |
| chr3_+_155860751 | 0.74 |
ENST00000471742.1
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr7_+_115850547 | 0.72 |
ENST00000358204.4
ENST00000455989.1 ENST00000537767.1 |
TES
|
testis derived transcript (3 LIM domains) |
| chr14_+_45553296 | 0.71 |
ENST00000355765.6
ENST00000553605.1 |
PRPF39
|
pre-mRNA processing factor 39 |
| chr15_+_32322709 | 0.71 |
ENST00000455693.2
|
CHRNA7
|
cholinergic receptor, nicotinic, alpha 7 (neuronal) |
| chr1_+_156338993 | 0.68 |
ENST00000368249.1
ENST00000368246.2 ENST00000537040.1 ENST00000400992.2 ENST00000255013.3 ENST00000451864.2 |
RHBG
|
Rh family, B glycoprotein (gene/pseudogene) |
| chr2_+_234216454 | 0.66 |
ENST00000447536.1
ENST00000409110.1 |
SAG
|
S-antigen; retina and pineal gland (arrestin) |
| chr15_+_80351910 | 0.65 |
ENST00000261749.6
ENST00000561060.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr2_-_128399706 | 0.60 |
ENST00000426981.1
|
LIMS2
|
LIM and senescent cell antigen-like domains 2 |
| chr1_-_26372602 | 0.55 |
ENST00000374278.3
ENST00000374276.3 |
SLC30A2
|
solute carrier family 30 (zinc transporter), member 2 |
| chrX_+_135730373 | 0.55 |
ENST00000370628.2
|
CD40LG
|
CD40 ligand |
| chr22_+_26565440 | 0.51 |
ENST00000404234.3
ENST00000529632.2 ENST00000360929.3 ENST00000248933.6 ENST00000343706.4 |
SEZ6L
|
seizure related 6 homolog (mouse)-like |
| chr20_+_35807512 | 0.50 |
ENST00000373622.5
|
RPN2
|
ribophorin II |
| chr17_+_55163075 | 0.49 |
ENST00000571629.1
ENST00000570423.1 ENST00000575186.1 ENST00000573085.1 ENST00000572814.1 |
AKAP1
|
A kinase (PRKA) anchor protein 1 |
| chr3_+_39851094 | 0.47 |
ENST00000302541.6
|
MYRIP
|
myosin VIIA and Rab interacting protein |
| chr2_+_32502952 | 0.44 |
ENST00000238831.4
|
YIPF4
|
Yip1 domain family, member 4 |
| chr21_-_38445011 | 0.41 |
ENST00000464265.1
ENST00000399102.1 |
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr11_+_64323098 | 0.41 |
ENST00000301891.4
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr1_-_16302565 | 0.41 |
ENST00000537142.1
ENST00000448462.2 |
ZBTB17
|
zinc finger and BTB domain containing 17 |
| chr21_-_34852304 | 0.36 |
ENST00000542230.2
|
TMEM50B
|
transmembrane protein 50B |
| chr19_-_50370799 | 0.35 |
ENST00000600910.1
ENST00000322344.3 ENST00000600573.1 |
PNKP
|
polynucleotide kinase 3'-phosphatase |
| chr11_+_118443098 | 0.32 |
ENST00000392859.3
ENST00000359415.4 ENST00000534182.2 ENST00000264028.4 |
ARCN1
|
archain 1 |
| chr16_+_57279248 | 0.30 |
ENST00000562023.1
ENST00000563234.1 |
ARL2BP
|
ADP-ribosylation factor-like 2 binding protein |
| chr8_+_124084899 | 0.28 |
ENST00000287380.1
ENST00000309336.3 ENST00000519418.1 ENST00000327098.5 ENST00000522420.1 ENST00000521676.1 ENST00000378080.2 |
TBC1D31
|
TBC1 domain family, member 31 |
| chr7_-_99679324 | 0.24 |
ENST00000292393.5
ENST00000413658.2 ENST00000412947.1 ENST00000441298.1 ENST00000449785.1 ENST00000299667.4 ENST00000424697.1 |
ZNF3
|
zinc finger protein 3 |
| chrX_-_64754611 | 0.22 |
ENST00000374807.5
ENST00000374811.3 ENST00000374804.5 ENST00000312391.8 |
LAS1L
|
LAS1-like (S. cerevisiae) |
| chr13_-_108870623 | 0.22 |
ENST00000405925.1
|
LIG4
|
ligase IV, DNA, ATP-dependent |
| chrX_+_135730297 | 0.16 |
ENST00000370629.2
|
CD40LG
|
CD40 ligand |
| chr8_-_101734170 | 0.13 |
ENST00000522387.1
ENST00000518196.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr18_+_56892724 | 0.10 |
ENST00000456142.3
ENST00000530323.1 |
GRP
|
gastrin-releasing peptide |
| chr15_-_82824843 | 0.07 |
ENST00000560826.1
ENST00000559187.1 ENST00000330339.7 |
RPS17
|
ribosomal protein S17 |
| chr1_+_169337172 | 0.06 |
ENST00000367807.3
ENST00000367808.3 ENST00000329281.2 ENST00000420531.1 |
BLZF1
|
basic leucine zipper nuclear factor 1 |
| chr12_-_121476750 | 0.04 |
ENST00000543677.1
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr20_+_35807449 | 0.04 |
ENST00000237530.6
|
RPN2
|
ribophorin II |
| chr1_+_3614591 | 0.00 |
ENST00000378290.4
|
TP73
|
tumor protein p73 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HESX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.6 | GO:0032888 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 1.6 | 6.4 | GO:1901895 | negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 1.4 | 4.1 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 1.2 | 3.7 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 1.2 | 8.6 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 1.0 | 147.0 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.9 | 2.8 | GO:2000417 | negative regulation of eosinophil migration(GO:2000417) |
| 0.7 | 3.0 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.7 | 2.1 | GO:2000538 | regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.7 | 3.9 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.6 | 3.8 | GO:0052565 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.6 | 1.9 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.6 | 107.3 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.6 | 4.5 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.5 | 2.5 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.5 | 3.3 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.5 | 1.4 | GO:0001694 | histamine biosynthetic process(GO:0001694) |
| 0.4 | 2.6 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.4 | 3.0 | GO:0034350 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.4 | 3.7 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.3 | 2.9 | GO:0034086 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.3 | 0.9 | GO:0021778 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.3 | 4.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 0.7 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.2 | 3.8 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.2 | 0.7 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.2 | 2.7 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.2 | 2.1 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.2 | 1.1 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 0.2 | 1.0 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.2 | 1.3 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 0.7 | GO:0050893 | sensory processing(GO:0050893) |
| 0.1 | 0.8 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.1 | 1.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 6.5 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.1 | 1.1 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 1.7 | GO:0009650 | UV protection(GO:0009650) |
| 0.1 | 0.5 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 4.9 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.1 | 1.7 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 | 2.1 | GO:0016246 | RNA interference(GO:0016246) |
| 0.1 | 1.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 31.1 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.1 | 0.3 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.1 | 2.6 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.1 | 0.7 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 1.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 1.5 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.1 | 3.1 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 1.3 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.1 | 2.2 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.1 | 1.3 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 0.6 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 | 1.7 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.1 | 1.0 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 3.2 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 0.7 | GO:1903817 | diaphragm development(GO:0060539) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.5 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 2.2 | GO:0042417 | dopamine metabolic process(GO:0042417) |
| 0.0 | 0.1 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.8 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 1.0 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.5 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 2.2 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 1.4 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 2.3 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 1.2 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 1.2 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.9 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 1.6 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 2.5 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 1.1 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.7 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 1.2 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.2 | GO:0000478 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) |
| 0.0 | 1.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 4.0 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 1.3 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.0 | 0.4 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.0 | 0.5 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 1.1 | GO:0000725 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.0 | 0.3 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.6 | GO:0055028 | cortical microtubule(GO:0055028) mitotic spindle astral microtubule(GO:0061673) |
| 1.0 | 3.1 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.9 | 6.4 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.6 | 31.1 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.4 | 4.9 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.4 | 73.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.4 | 3.2 | GO:0097433 | dense body(GO:0097433) |
| 0.3 | 2.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.3 | 2.5 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.2 | 1.6 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.2 | 1.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.2 | 1.3 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.2 | 1.7 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.2 | 3.8 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 1.5 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 8.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 2.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 4.5 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 1.1 | GO:0061202 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.1 | 4.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 123.9 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 0.7 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 3.3 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.7 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 3.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.2 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 2.2 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 5.9 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 2.0 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 1.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 1.1 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 1.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 2.0 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 2.1 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 2.5 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.7 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 1.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.9 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.9 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 274.3 | GO:0003823 | antigen binding(GO:0003823) |
| 1.0 | 3.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.7 | 2.2 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.7 | 2.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.6 | 2.5 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.6 | 3.0 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.6 | 2.2 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.5 | 3.5 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.4 | 1.7 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.4 | 5.6 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.4 | 3.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.3 | 2.8 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.3 | 4.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.3 | 2.3 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.3 | 1.7 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.3 | 4.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.2 | 0.7 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.2 | 0.7 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.2 | 1.7 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.2 | 1.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.2 | 2.2 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.2 | 4.5 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 3.8 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 1.5 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.7 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 1.2 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 3.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 1.3 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.1 | 0.7 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 4.0 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.1 | 1.9 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 1.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 1.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 1.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 0.5 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.7 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 2.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 2.6 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 1.0 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 3.9 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.9 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 1.0 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 8.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.7 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 4.9 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.7 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.2 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.5 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 1.3 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 2.9 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.4 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 4.7 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 4.7 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 1.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.8 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 8.6 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 5.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 4.9 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 2.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 5.1 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 5.3 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.1 | 5.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 6.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 4.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 3.8 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 1.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 1.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.7 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.6 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.3 | 2.8 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.2 | 3.7 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 3.1 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 4.9 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 1.9 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 6.9 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 3.3 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 1.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.7 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 5.1 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 2.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 2.6 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 3.0 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 2.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 1.7 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 1.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 3.0 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.8 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 3.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 4.2 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 4.0 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.7 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.1 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 2.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 4.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 2.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 3.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.3 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.2 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 1.1 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |