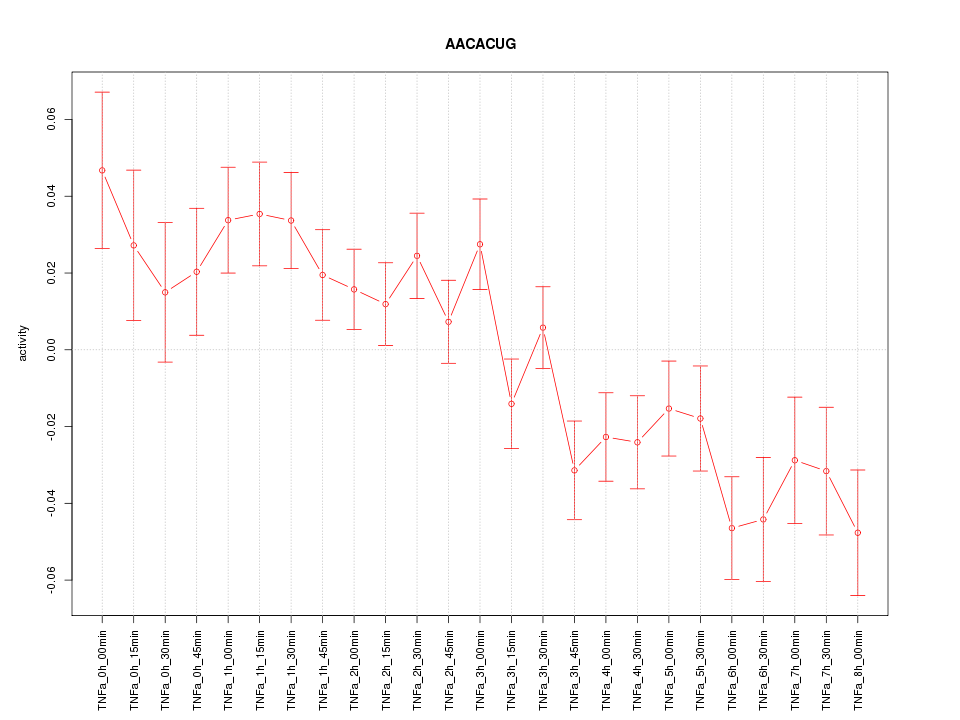
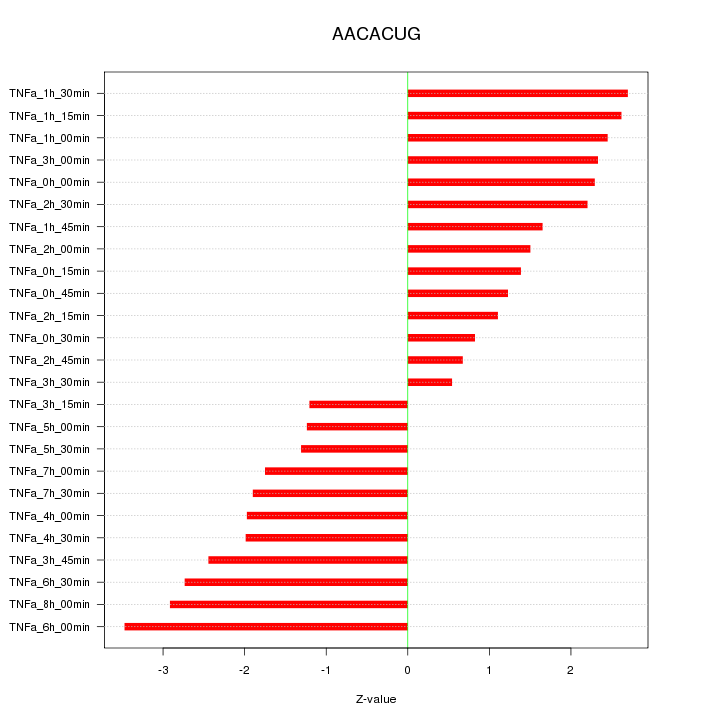
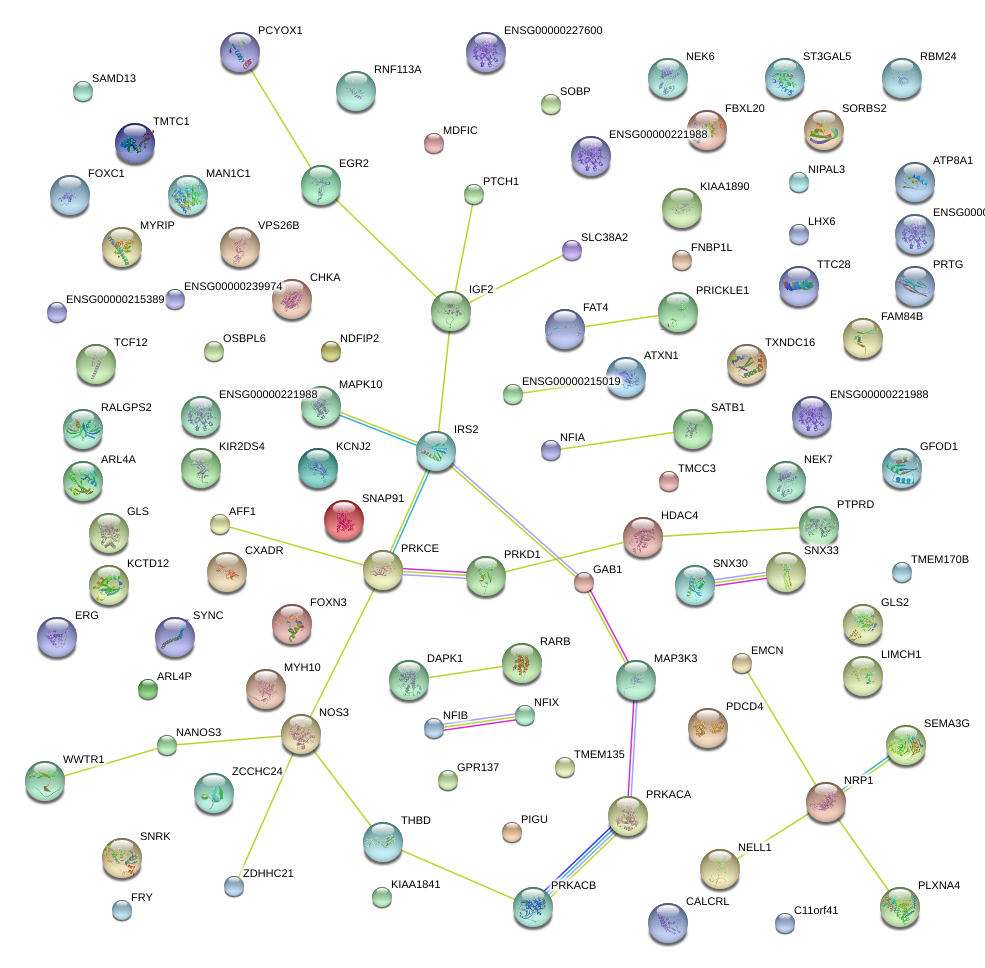
|
chr1_+_61542928
|
6.958
|
NM_001145511
|
NFIA
|
nuclear factor I/A
|
|
chr1_+_61547625
|
6.637
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr1_+_61547388
|
6.224
|
NM_001145512
|
NFIA
|
nuclear factor I/A
|
|
chr4_-_186697065
|
6.133
|
NM_001145673
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr10_-_81205091
|
6.038
|
NM_153367
|
ZCCHC24
|
zinc finger, CCHC domain containing 24
|
|
chr4_-_186733372
|
6.032
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr4_-_186732047
|
5.928
|
NM_001145672
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr4_-_186732208
|
5.805
|
NM_001145671
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr4_-_186877869
|
5.686
|
NM_021069
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr1_+_84630575
|
5.681
|
NM_001242860
NM_001242861
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr1_+_84609941
|
5.635
|
NM_182948
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr1_+_84630014
|
5.572
|
NM_001242857
NM_001242858
NM_001242859
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr17_-_8534008
|
5.347
|
NM_005964
|
MYH10
|
myosin, heavy chain 10, non-muscle
|
|
chr1_+_84647342
|
5.230
|
NM_001242862
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr10_+_112631552
|
4.842
|
NM_001199492
NM_014456
NM_145341
|
PDCD4
|
programmed cell death 4 (neoplastic transformation inhibitor)
|
|
chr4_+_41362750
|
4.826
|
NM_001112717
NM_001112718
NM_014988
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr6_+_11538486
|
4.773
|
NM_001100829
|
TMEM170B
|
transmembrane protein 170B
|
|
chr7_+_12726413
|
4.649
|
NM_001037164
NM_212460
|
ARL4A
|
ADP-ribosylation factor-like 4A
|
|
chr4_+_41614911
|
4.644
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr6_-_16761600
|
4.544
|
NM_000332
NM_001128164
|
ATXN1
|
ataxin 1
|
|
chr7_+_12726910
|
4.520
|
NM_001195396
NM_005738
|
ARL4A
|
ADP-ribosylation factor-like 4A
|
|
chr13_-_77460432
|
4.485
|
NM_138444
|
KCTD12
|
potassium channel tetramerisation domain containing 12
|
|
chr9_-_10612722
|
4.285
|
NM_002839
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr9_-_8733893
|
4.142
|
NM_001040712
NM_001171025
NM_130391
NM_130392
NM_130393
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr2_+_191745546
|
3.941
|
NM_014905
|
GLS
|
glutaminase
|
|
chr4_-_186578122
|
3.916
|
NM_001145674
NM_001145675
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr6_-_84419108
|
3.819
|
NM_001242793
NM_001242794
NM_014841
|
SNAP91
|
synaptosomal-associated protein, 91kDa homolog (mouse)
|
|
chr8_-_127570710
|
3.737
|
NM_174911
|
FAM84B
|
family with sequence similarity 84, member B
|
|
chr9_+_127054246
|
3.697
|
NM_001166170
NM_001166171
|
NEK6
|
NIMA (never in mitosis gene a)-related kinase 6
|
|
chr6_-_84418705
|
3.671
|
NM_001242792
|
SNAP91
|
synaptosomal-associated protein, 91kDa homolog (mouse)
|
|
chr2_-_240322620
|
3.638
|
NM_006037
|
HDAC4
|
histone deacetylase 4
|
|
chr1_+_25943958
|
3.589
|
NM_020379
|
MAN1C1
|
mannosidase, alpha, class 1C, member 1
|
|
chr3_+_39851028
|
3.562
|
NM_015460
|
MYRIP
|
myosin VIIA and Rab interacting protein
|
|
chr4_-_42658883
|
3.560
|
NM_001105529
NM_006095
|
ATP8A1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1
|
|
chr9_+_127054567
|
3.532
|
NM_001166169
|
NEK6
|
NIMA (never in mitosis gene a)-related kinase 6
|
|
chr21_+_18885104
|
3.516
|
NM_001207063
NM_001207064
NM_001207065
NM_001207066
NM_001338
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chr3_+_25469833
|
3.512
|
NM_000965
|
RARB
|
retinoic acid receptor, beta
|
|
chr3_-_149375584
|
3.308
|
NM_001168280
NM_015472
|
WWTR1
|
WW domain containing transcription regulator 1
|
|
chr3_-_149421059
|
3.162
|
NM_001168278
|
WWTR1
|
WW domain containing transcription regulator 1
|
|
chr3_+_43328001
|
3.077
|
NM_001100594
NM_017719
|
SNRK
|
SNF related kinase
|
|
chr13_+_32605436
|
3.061
|
NM_023037
|
FRY
|
furry homolog (Drosophila)
|
|
chr17_+_68165660
|
3.047
|
NM_000891
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2
|
|
chr2_-_86094792
|
3.022
|
NM_001042437
|
ST3GAL5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5
|
|
chr9_+_127019782
|
2.981
|
NM_001166167
NM_001145001
NM_014397
|
NEK6
|
NIMA (never in mitosis gene a)-related kinase 6
|
|
chr13_-_110438896
|
2.935
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr9_+_127020462
|
2.869
|
NM_001166168
|
NEK6
|
NIMA (never in mitosis gene a)-related kinase 6
|
|
chr11_+_86748870
|
2.836
|
NM_001168724
NM_022918
|
TMEM135
|
transmembrane protein 135
|
|
chr14_-_89883306
|
2.822
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr4_+_144257982
|
2.783
|
NM_002039
NM_207123
|
GAB1
|
GRB2-associated binding protein 1
|
|
chr7_-_132261268
|
2.707
|
NM_001105543
NM_020911
|
PLXNA4
|
plexin A4
|
|
chr12_-_29937691
|
2.660
|
NM_175861
|
TMTC1
|
transmembrane and tetratricopeptide repeat containing 1
|
|
chr12_-_29936685
|
2.641
|
NM_001193451
|
TMTC1
|
transmembrane and tetratricopeptide repeat containing 1
|
|
chr1_+_178694273
|
2.599
|
NM_152663
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2
|
|
chr11_-_2162340
|
2.576
|
NM_001127598
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr2_-_86116016
|
2.563
|
NM_003896
|
ST3GAL5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5
|
|
chr11_+_33563772
|
2.455
|
NM_012194
|
C11orf41
|
chromosome 11 open reading frame 41
|
|
chr8_-_4852262
|
2.436
|
NM_033225
|
CSMD1
|
CUB and Sushi multiple domains 1
|
|
chr6_+_107811272
|
2.377
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr11_-_2170792
|
2.374
|
NM_001007139
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr14_-_90085325
|
2.333
|
NM_001085471
|
FOXN3
|
forkhead box N3
|
|
chr11_+_134094545
|
2.322
|
NM_052875
|
VPS26B
|
vacuolar protein sorting 26 homolog B (S. pombe)
|
|
chr2_+_45878912
|
2.270
|
NM_005400
|
PRKCE
|
protein kinase C, epsilon
|
|
chr11_-_2160199
|
2.225
|
NM_000612
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr6_+_17282931
|
2.186
|
NM_153020
|
RBM24
|
RNA binding motif protein 24
|
|
chr20_-_23030141
|
2.169
|
NM_000361
|
THBD
|
thrombomodulin
|
|
chr4_+_126237566
|
2.147
|
NM_024582
|
FAT4
|
FAT tumor suppressor homolog 4 (Drosophila)
|
|
chr9_-_14693459
|
2.133
|
NM_178566
|
ZDHHC21
|
zinc finger, DHHC-type containing 21
|
|
chr9_+_90112754
|
2.075
|
NM_004938
|
DAPK1
|
death-associated protein kinase 1
|
|
chr22_-_29075708
|
2.062
|
NM_001145418
|
TTC28
|
tetratricopeptide repeat domain 28
|
|
chr9_-_124984018
|
2.051
|
NM_001242335
|
LHX6
|
LIM homeobox 6
|
|
chr13_+_80055257
|
2.047
|
NM_001161407
NM_019080
|
NDFIP2
|
Nedd4 family interacting protein 2
|
|
chr12_-_95044205
|
1.997
|
NM_020698
|
TMCC3
|
transmembrane and coiled-coil domain family 3
|
|
chr1_+_93913566
|
1.980
|
NM_001024948
NM_001164473
NM_017737
|
FNBP1L
|
formin binding protein 1-like
|
|
chr6_+_1610680
|
1.947
|
NM_001453
|
FOXC1
|
forkhead box C1
|
|
chr12_-_42877785
|
1.927
|
NM_001144882
NM_001144883
|
PRICKLE1
|
prickle homolog 1 (Drosophila)
|
|
chr12_-_42877413
|
1.898
|
NM_001144881
|
PRICKLE1
|
prickle homolog 1 (Drosophila)
|
|
chr6_+_17282294
|
1.872
|
NM_001143941
|
RBM24
|
RNA binding motif protein 24
|
|
chr6_+_17281773
|
1.853
|
NM_001143942
|
RBM24
|
RNA binding motif protein 24
|
|
chr12_-_42983412
|
1.851
|
NM_153026
|
PRICKLE1
|
prickle homolog 1 (Drosophila)
|
|
chr12_-_46766563
|
1.849
|
NM_018976
|
SLC38A2
|
solute carrier family 38, member 2
|
|
chr17_-_37557851
|
1.823
|
NM_001184906
NM_032875
|
FBXL20
|
F-box and leucine-rich repeat protein 20
|
|
chr10_-_64578926
|
1.822
|
NM_001136178
|
EGR2
|
early growth response 2
|
|
chr9_+_115513050
|
1.817
|
NM_001012994
|
SNX30
|
sorting nexin family member 30
|
|
chr9_-_124990706
|
1.811
|
NM_001242333
|
LHX6
|
LIM homeobox 6
|
|
chr15_-_56035176
|
1.798
|
NM_173814
|
PRTG
|
protogenin
|
|
chr4_-_101439071
|
1.786
|
NM_001159694
NM_016242
|
EMCN
|
endomucin
|
|
chr10_-_33623771
|
1.782
|
NM_001024628
NM_001024629
NM_001244972
NM_001244973
NM_003873
|
NRP1
|
neuropilin 1
|
|
chr3_-_18466759
|
1.725
|
NM_001195470
NM_002971
|
SATB1
|
SATB homeobox 1
|
|
chr10_-_64576123
|
1.724
|
NM_000399
NM_001136177
NM_001136179
|
EGR2
|
early growth response 2
|
|
chr2_+_179184970
|
1.721
|
NM_145739
|
OSBPL6
|
oxysterol binding protein-like 6
|
|
chr3_-_18480203
|
1.717
|
NM_001131010
|
SATB1
|
SATB homeobox 1
|
|
chr2_+_179059071
|
1.702
|
NM_001201480
NM_001201481
NM_001201482
NM_032523
|
OSBPL6
|
oxysterol binding protein-like 6
|
|
chr1_+_24742244
|
1.699
|
NM_020448
|
NIPAL3
|
NIPA-like domain containing 3
|
|
chr11_-_67888857
|
1.699
|
NM_001277
NM_212469
|
CHKA
|
choline kinase alpha
|
|
chr2_+_70485204
|
1.696
|
NM_016297
|
PCYOX1
|
prenylcysteine oxidase 1
|
|
chr2_+_61292999
|
1.683
|
NM_001129993
|
KIAA1841
|
KIAA1841
|
|
chr9_-_98270321
|
1.681
|
NM_001083604
NM_001083605
|
PTCH1
|
patched 1
|
|
chr21_-_40033617
|
1.668
|
NM_001243432
NM_001136154
NM_004449
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr6_-_13408368
|
1.659
|
NM_001242630
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1
|
|
chr7_+_114562140
|
1.656
|
NM_001166345
NM_001166346
NM_199072
|
MDFIC
|
MyoD family inhibitor domain containing
|
|
chr9_-_98269480
|
1.655
|
NM_001083607
|
PTCH1
|
patched 1
|
|
chr2_-_188312872
|
1.649
|
NM_005795
|
CALCRL
|
calcitonin receptor-like
|
|
chr7_+_150690838
|
1.649
|
NM_001160109
NM_001160110
NM_001160111
|
NOS3
|
nitric oxide synthase 3 (endothelial cell)
|
|
chr21_-_39870303
|
1.648
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr1_-_33168356
|
1.645
|
NM_001161708
NM_030786
|
SYNC
|
syncoilin, intermediate filament protein
|
|
chr4_+_87928083
|
1.639
|
NM_005935
|
AFF1
|
AF4/FMR2 family, member 1
|
|
chr1_+_84764048
|
1.639
|
NM_001010971
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr14_-_30396811
|
1.630
|
NM_002742
|
PRKD1
|
protein kinase D1
|
|
chr1_+_84768333
|
1.615
|
NM_001134664
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr3_-_52479042
|
1.611
|
NM_020163
|
SEMA3G
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3G
|
|
chr17_+_61699786
|
1.608
|
NM_002401
NM_203351
|
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3
|
|
chr1_+_84767288
|
1.596
|
NM_001134663
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr6_+_32121228
|
1.573
|
NM_138717
|
PPT2
|
palmitoyl-protein thioesterase 2
|
|
chr9_-_124990949
|
1.554
|
NM_014368
NM_199160
|
LHX6
|
LIM homeobox 6
|
|
chr15_+_57210823
|
1.518
|
NM_003205
NM_207036
NM_207037
NM_207038
|
TCF12
|
transcription factor 12
|
|
chr9_-_98270830
|
1.504
|
NM_000264
|
PTCH1
|
patched 1
|
|
chr2_+_178257371
|
1.501
|
NM_003659
|
AGPS
|
alkylglycerone phosphate synthase
|
|
chr13_-_74707913
|
1.501
|
NM_007249
|
KLF12
|
Kruppel-like factor 12
|
|
chr14_+_53019865
|
1.486
|
NM_001099652
|
GPR137C
|
G protein-coupled receptor 137C
|
|
chr4_-_85887543
|
1.470
|
NM_014991
|
WDFY3
|
WD repeat and FYVE domain containing 3
|
|
chr1_-_154474524
|
1.466
|
NM_001010846
|
SHE
|
Src homology 2 domain containing E
|
|
chr6_+_17600504
|
1.451
|
NM_016255
|
FAM8A1
|
family with sequence similarity 8, member A1
|
|
chr6_-_52441815
|
1.447
|
NM_012288
|
TRAM2
|
translocation associated membrane protein 2
|
|
chr12_-_56615679
|
1.446
|
NM_005785
NM_194358
NM_194359
|
RNF41
|
ring finger protein 41
|
|
chr1_+_82266081
|
1.432
|
NM_012302
|
LPHN2
|
latrophilin 2
|
|
chr5_+_95066628
|
1.415
|
NM_014899
|
RHOBTB3
|
Rho-related BTB domain containing 3
|
|
chr8_+_97505876
|
1.413
|
NM_002998
|
SDC2
|
syndecan 2
|
|
chr12_-_56615484
|
1.409
|
NM_001242826
|
RNF41
|
ring finger protein 41
|
|
chr19_-_31840118
|
1.408
|
NM_020856
|
TSHZ3
|
teashirt zinc finger homeobox 3
|
|
chr2_+_203241044
|
1.406
|
NM_001204
|
BMPR2
|
bone morphogenetic protein receptor, type II (serine/threonine kinase)
|
|
chr3_-_141719188
|
1.403
|
NM_001178140
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr7_+_17338182
|
1.389
|
NM_001621
|
AHR
|
aryl hydrocarbon receptor
|
|
chr10_-_126849556
|
1.382
|
NM_001083914
|
CTBP2
|
C-terminal binding protein 2
|
|
chr3_+_32148000
|
1.369
|
NM_015141
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like
|
|
chr5_+_153570270
|
1.366
|
NM_198321
|
GALNT10
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10)
|
|
chr18_+_8717368
|
1.356
|
NM_015210
|
CCDC165
|
coiled-coil domain containing 165
|
|
chr9_-_79520878
|
1.344
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr1_+_52082545
|
1.339
|
NM_024586
NM_148906
NM_148908
NM_148909
|
OSBPL9
|
oxysterol binding protein-like 9
|
|
chr1_+_52195458
|
1.337
|
NM_148904
NM_148905
NM_148907
|
OSBPL9
|
oxysterol binding protein-like 9
|
|
chr7_-_105319608
|
1.333
|
NM_138495
|
ATXN7L1
|
ataxin 7-like 1
|
|
chr12_+_27397077
|
1.331
|
NM_015000
|
STK38L
|
serine/threonine kinase 38 like
|
|
chr11_+_92085261
|
1.325
|
NM_001008781
|
FAT3
|
FAT tumor suppressor homolog 3 (Drosophila)
|
|
chr6_-_35696359
|
1.290
|
NM_001145775
|
FKBP5
|
FK506 binding protein 5
|
|
chr1_-_225616556
|
1.283
|
NM_194442
|
LBR
|
lamin B receptor
|
|
chr15_+_57511616
|
1.276
|
NM_207040
|
TCF12
|
transcription factor 12
|
|
chr3_-_149051258
|
1.275
|
NM_001184723
NM_138786
|
TM4SF18
|
transmembrane 4 L six family member 18
|
|
chrX_+_13671224
|
1.263
|
NM_152634
|
TCEANC
|
transcription elongation factor A (SII) N-terminal and central domain containing
|
|
chr18_+_13611664
|
1.257
|
NM_001003674
NM_001003675
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr3_-_17741511
|
1.255
|
NM_001134381
|
TBC1D5
|
TBC1 domain family, member 5
|
|
chr20_-_23066828
|
1.246
|
NM_012072
|
CD93
|
CD93 molecule
|
|
chr6_+_32121775
|
1.226
|
NM_001204103
NM_005155
|
PPT2-EGFL8
PPT2
|
PPT2-EGFL8 readthrough
palmitoyl-protein thioesterase 2
|
|
chr6_+_116832807
|
1.214
|
NM_153711
|
FAM26E
|
family with sequence similarity 26, member E
|
|
chr15_+_63569748
|
1.212
|
NM_001145646
NM_031301
|
APH1B
|
anterior pharynx defective 1 homolog B (C. elegans)
|
|
chrX_+_21857655
|
1.196
|
NM_015884
|
MBTPS2
|
membrane-bound transcription factor peptidase, site 2
|
|
chr7_-_11871735
|
1.185
|
NM_015204
|
THSD7A
|
thrombospondin, type I, domain containing 7A
|
|
chr17_-_49337370
|
1.172
|
NM_017643
|
MBTD1
|
mbt domain containing 1
|
|
chr10_-_126716452
|
1.171
|
NM_022802
|
CTBP2
|
C-terminal binding protein 2
|
|
chr12_+_50898744
|
1.169
|
NM_173602
|
DIP2B
|
DIP2 disco-interacting protein 2 homolog B (Drosophila)
|
|
chr11_-_63536112
|
1.168
|
NM_001144936
|
C11orf95
|
chromosome 11 open reading frame 95
|
|
chr18_+_13620866
|
1.165
|
NM_004338
NM_181483
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr9_+_5450455
|
1.142
|
NM_014143
|
CD274
|
CD274 molecule
|
|
chr1_-_179112178
|
1.140
|
NM_001136000
NM_001168239
NM_005158
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr8_-_38326351
|
1.140
|
NM_001174063
NM_001174064
NM_015850
NM_023105
NM_023106
NM_023110
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr4_+_87856152
|
1.138
|
NM_001166693
|
AFF1
|
AF4/FMR2 family, member 1
|
|
chr9_-_16870719
|
1.133
|
NM_017637
|
BNC2
|
basonuclin 2
|
|
chr18_-_53255734
|
1.129
|
NM_001083962
NM_001243230
NM_003199
|
TCF4
|
transcription factor 4
|
|
chr1_+_114522029
|
1.128
|
NM_020190
|
OLFML3
|
olfactomedin-like 3
|
|
chr8_-_81787015
|
1.124
|
NM_001033723
|
ZNF704
|
zinc finger protein 704
|
|
chr9_+_131644780
|
1.121
|
NM_001127244
|
LRRC8A
|
leucine rich repeat containing 8 family, member A
|
|
chr1_+_84543629
|
1.108
|
NM_002731
NM_207578
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr11_+_117049838
|
1.105
|
NM_001040455
|
SIDT2
|
SID1 transmembrane family, member 2
|
|
chr4_+_55523906
|
1.098
|
NM_000222
NM_001093772
|
KIT
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog
|
|
chr8_-_38325241
|
1.097
|
NM_001174065
NM_001174066
NM_001174067
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr5_-_111093251
|
1.093
|
NM_001142479
NM_001142480
NM_001142478
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr3_-_17782398
|
1.088
|
NM_014744
|
TBC1D5
|
TBC1 domain family, member 5
|
|
chr10_-_126849066
|
1.083
|
NM_001329
|
CTBP2
|
C-terminal binding protein 2
|
|
chrX_-_119709618
|
1.077
|
NM_003588
|
CUL4B
|
cullin 4B
|
|
chr19_-_48752921
|
1.074
|
NM_001184901
NM_001184903
NM_014959
|
CARD8
|
caspase recruitment domain family, member 8
|
|
chr18_+_59992519
|
1.070
|
NM_003839
|
TNFRSF11A
|
tumor necrosis factor receptor superfamily, member 11a, NFKB activator
|
|
chr10_+_65281108
|
1.067
|
NM_001001330
|
REEP3
|
receptor accessory protein 3
|
|
chr3_-_64211111
|
1.067
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr7_-_35077652
|
1.064
|
NM_015283
|
DPY19L1
|
dpy-19-like 1 (C. elegans)
|
|
chr10_+_102106771
|
1.063
|
NM_005063
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase)
|
|
chr17_+_36584692
|
1.060
|
NM_001199417
|
ARHGAP23
|
Rho GTPase activating protein 23
|
|
chr5_-_88179278
|
1.058
|
NM_001193350
NM_002397
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr19_-_48744276
|
1.053
|
NM_001184900
|
CARD8
|
caspase recruitment domain family, member 8
|
|
chr4_-_129209983
|
1.052
|
NM_006320
|
PGRMC2
|
progesterone receptor membrane component 2
|
|
chr17_-_74137370
|
1.051
|
NM_001454
|
FOXJ1
|
forkhead box J1
|
|
chr12_+_12938540
|
1.051
|
NM_030817
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr7_+_100136784
|
1.050
|
NM_006076
|
AGFG2
|
ArfGAP with FG repeats 2
|
|
chr1_-_225840660
|
1.048
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr8_-_17555158
|
1.048
|
NM_020749
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr5_-_111093030
|
1.045
|
NM_001142482
NM_001142481
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr3_-_56502364
|
1.035
|
NM_015576
|
ERC2
|
ELKS/RAB6-interacting/CAST family member 2
|
|
chr11_+_74459900
|
1.031
|
NM_001098638
|
RNF169
|
ring finger protein 169
|
|
chr11_+_134123406
|
1.030
|
NM_014384
|
ACAD8
|
acyl-CoA dehydrogenase family, member 8
|
|
chr9_+_131644368
|
1.023
|
NM_001127245
NM_019594
|
LRRC8A
|
leucine rich repeat containing 8 family, member A
|
|
chr3_-_17784096
|
1.018
|
NM_001134380
|
TBC1D5
|
TBC1 domain family, member 5
|
|
chr1_-_92371558
|
1.011
|
NM_001195684
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr1_+_109102970
|
1.000
|
NM_001010883
|
FAM102B
|
family with sequence similarity 102, member B
|