|
chr18_+_42260774
|
0.256
|
NM_015559
|
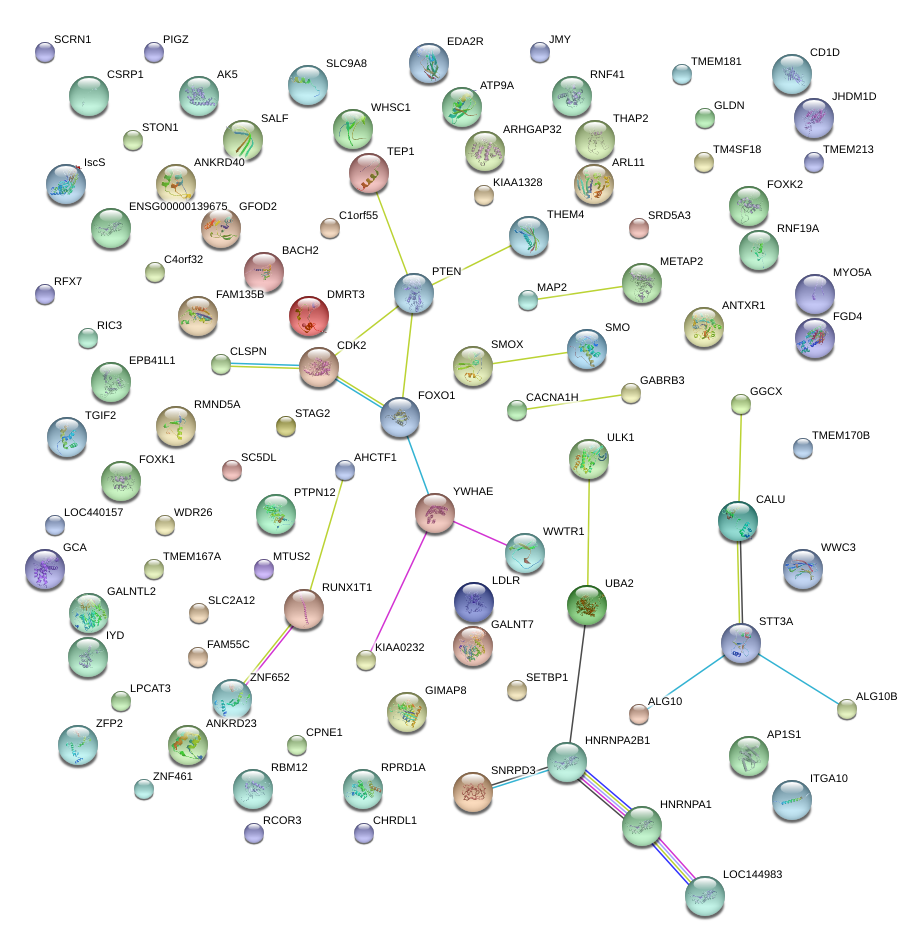
SETBP1
|
SET binding protein 1
|
|
chr8_-_93029864
|
0.217
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_-_93107881
|
0.199
|
NM_001198625
NM_001198626
NM_001198627
NM_001198631
NM_001198634
NM_001198679
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_-_93111915
|
0.191
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_-_93107442
|
0.190
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_-_93115453
|
0.147
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_-_93075190
|
0.145
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr3_-_149051258
|
0.130
|
NM_001184723
NM_138786
|
TM4SF18
|
transmembrane 4 L six family member 18
|
|
chr6_+_11538486
|
0.128
|
NM_001100829
|
TMEM170B
|
transmembrane protein 170B
|
|
chrX_+_9983793
|
0.125
|
NM_015691
|
WWC3
|
WWC family member 3
|
|
chr3_-_149375584
|
0.123
|
NM_001168280
NM_015472
|
WWTR1
|
WW domain containing transcription regulator 1
|
|
chr3_-_149421059
|
0.120
|
NM_001168278
|
WWTR1
|
WW domain containing transcription regulator 1
|
|
chr7_-_30029904
|
0.096
|
NM_001145514
|
SCRN1
|
secernin 1
|
|
chr7_-_30029284
|
0.087
|
NM_001145515
NM_014766
NM_001145513
|
SCRN1
|
secernin 1
|
|
chr9_+_976963
|
0.086
|
NM_021240
|
DMRT3
|
doublesex and mab-3 related transcription factor 3
|
|
chr2_+_48757292
|
0.071
|
NM_001198595
NM_006873
|
STON1
|
stonin 1
|
|
chr1_+_77748286
|
0.071
|
NM_012093
|
AK5
|
adenylate kinase 5
|
|
chr1_+_77747602
|
0.071
|
NM_174858
|
AK5
|
adenylate kinase 5
|
|
chr7_+_150147936
|
0.070
|
NM_175571
|
GIMAP8
|
GTPase, IMAP family member 8
|
|
chrX_+_123095555
|
0.068
|
NM_001042750
NM_001042751
NM_006603
|
STAG2
|
stromal antigen 2
|
|
chr1_+_145524989
|
0.064
|
NM_003637
|
ITGA10
|
integrin, alpha 10
|
|
chr6_-_91006580
|
0.061
|
NM_001170794
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr20_+_34742656
|
0.058
|
NM_012156
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr16_+_1203240
|
0.054
|
NM_001005407
NM_021098
|
CACNA1H
|
calcium channel, voltage-dependent, T type, alpha 1H subunit
|
|
chr20_+_34700257
|
0.054
|
NM_177996
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr7_-_26240366
|
0.053
|
NM_002137
NM_031243
|
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1
|
|
chrX_+_123094377
|
0.052
|
NM_001042749
|
STAG2
|
stromal antigen 2
|
|
chr6_-_91006460
|
0.049
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr12_+_56360547
|
0.046
|
NM_001798
NM_052827
|
CDK2
|
cyclin-dependent kinase 2
|
|
chr13_+_30002776
|
0.043
|
NM_015233
|
MTUS2
|
microtubule associated tumor suppressor candidate 2
|
|
chr12_+_32654976
|
0.041
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr19_+_11200037
|
0.038
|
NM_000527
NM_001195798
NM_001195799
NM_001195800
NM_001195802
NM_001195803
|
LDLR
|
low density lipoprotein receptor
|
|
chr2_+_86947387
|
0.037
|
NM_022780
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae)
|
|
chr7_+_77166750
|
0.037
|
NM_002835
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12
|
|
chr4_+_174089889
|
0.037
|
NM_017423
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7)
|
|
chr20_+_35201854
|
0.036
|
NM_001199513
NM_001199514
|
TGIF2
|
TGFB-induced factor homeobox 2
|
|
chr8_-_101322245
|
0.035
|
NM_183419
|
RNF19A
|
ring finger protein 19A
|
|
chr15_-_27018917
|
0.034
|
NM_021912
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3
|
|
chr20_+_35201948
|
0.034
|
NM_021809
|
TGIF2
|
TGFB-induced factor homeobox 2
|
|
chr8_-_101315457
|
0.033
|
NM_015435
|
RNF19A
|
ring finger protein 19A
|
|
chr11_+_125462736
|
0.033
|
NM_152713
|
STT3A
|
STT3, subunit of the oligosaccharyltransferase complex, homolog A (S. cerevisiae)
|
|
chr15_-_26961911
|
0.032
|
NM_001191320
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3
|
|
chr7_+_77167351
|
0.031
|
NM_001131008
NM_001131009
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12
|
|
chr12_-_56615679
|
0.030
|
NM_005785
NM_194358
NM_194359
|
RNF41
|
ring finger protein 41
|
|
chr12_-_56615484
|
0.030
|
NM_001242826
|
RNF41
|
ring finger protein 41
|
|
chr6_+_158957467
|
0.029
|
NM_020823
|
TMEM181
|
transmembrane protein 181
|
|
chr11_-_8190535
|
0.028
|
NM_001135109
NM_001206671
NM_001206672
NM_024557
|
RIC3
|
resistance to inhibitors of cholinesterase 3 homolog (C. elegans)
|
|
chr19_-_37157718
|
0.028
|
NM_153257
|
ZNF461
|
zinc finger protein 461
|
|
chr1_-_151882360
|
0.027
|
NM_053055
|
THEM4
|
thioesterase superfamily member 4
|
|
chr13_+_29598746
|
0.026
|
NM_001033602
|
MTUS2
|
microtubule associated tumor suppressor candidate 2
|
|
chr20_-_34252861
|
0.026
|
NM_001198838
NM_001198840
NM_006047
NM_152838
NM_001198863
NM_152925
NM_152926
NM_152927
NM_152928
|
RBM12
CPNE1
|
RNA binding motif protein 12
copine I
|
|
chr3_+_101498285
|
0.026
|
NM_001134456
|
FAM55C
|
family with sequence similarity 55, member C
|
|
chr5_-_82373253
|
0.024
|
NM_174909
|
TMEM167A
|
transmembrane protein 167A
|
|
chr19_+_34919213
|
0.022
|
NM_005499
|
UBA2
|
ubiquitin-like modifier activating enzyme 2
|
|
chr20_-_50384899
|
0.022
|
NM_006045
|
ATP9A
|
ATPase, class II, type 9A
|
|
chr1_-_226187055
|
0.022
|
NM_152608
|
C1orf55
|
chromosome 1 open reading frame 55
|
|
chr18_-_33647336
|
0.022
|
NM_018170
|
RPRD1A
|
regulation of nuclear pre-mRNA domain containing 1A
|
|
chr6_-_20212628
|
0.020
|
NM_001080480
|
MBOAT1
|
membrane bound O-acyltransferase domain containing 1
|
|
chr3_+_101498028
|
0.020
|
NM_145037
|
FAM55C
|
family with sequence similarity 55, member C
|
|
chr2_+_69240137
|
0.018
|
NM_018153
NM_032208
NM_053034
|
ANTXR1
|
anthrax toxin receptor 1
|
|
chr1_+_211433285
|
0.018
|
NM_018254
|
RCOR3
|
REST corepressor 3
|
|
chr11_+_121163501
|
0.018
|
NM_001024956
|
SC5DL
|
sterol-C5-desaturase (ERG3 delta-5-desaturase homolog, S. cerevisiae)-like
|
|
chr22_+_24951617
|
0.017
|
NM_004175
|
SNRPD3
|
small nuclear ribonucleoprotein D3 polypeptide 18kDa
|
|
chr12_+_132379262
|
0.017
|
NM_003565
|
ULK1
|
unc-51-like kinase 1 (C. elegans)
|
|
chr11_+_121163343
|
0.016
|
NM_006918
|
SC5DL
|
sterol-C5-desaturase (ERG3 delta-5-desaturase homolog, S. cerevisiae)-like
|
|
chr7_-_139876718
|
0.016
|
NM_030647
|
JHDM1D
|
jumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae)
|
|
chr12_+_72057676
|
0.016
|
NM_031435
|
THAP2
|
THAP domain containing, apoptosis associated protein 2
|
|
chr18_+_34409058
|
0.016
|
NM_020776
|
KIAA1328
|
KIAA1328
|
|
chr17_-_47439326
|
0.015
|
NM_001145365
NM_014897
|
ZNF652
|
zinc finger protein 652
|
|
chrX_-_65859107
|
0.015
|
NM_001199687
|
EDA2R
|
ectodysplasin A2 receptor
|
|
chr13_-_41240607
|
0.015
|
NM_002015
|
FOXO1
|
forkhead box O1
|
|
chr5_+_78531848
|
0.015
|
NM_152405
|
JMY
|
junction mediating and regulatory protein, p53 cofactor
|
|
chr11_-_128894008
|
0.014
|
NM_014715
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr1_+_220921576
|
0.014
|
NM_017898
|
MARC2
|
mitochondrial amidoxime reducing component 2
|
|
chr1_-_36235422
|
0.014
|
NM_001190481
NM_022111
|
CLSPN
|
claspin
|
|
chr7_+_128379345
|
0.014
|
NM_001130674
NM_001199671
NM_001199672
NM_001199673
NM_001199674
NM_001219
|
CALU
|
calumenin
|
|
chrX_-_110039285
|
0.013
|
NM_001143981
NM_001143982
NM_001143983
|
CHRDL1
|
chordin-like 1
|
|
chr2_+_210288668
|
0.013
|
NM_001039538
|
MAP2
|
microtubule-associated protein 2
|
|
chrX_-_65835845
|
0.013
|
NM_001242310
|
EDA2R
|
ectodysplasin A2 receptor
|
|
chr7_+_128828712
|
0.013
|
NM_005631
|
SMO
|
smoothened, frizzled family receptor
|
|
chrX_-_65858882
|
0.013
|
NM_021783
|
EDA2R
|
ectodysplasin A2 receptor
|
|
chr17_-_48785016
|
0.013
|
NM_052855
|
ANKRD40
|
ankyrin repeat domain 40
|
|
chr15_-_52821004
|
0.013
|
NM_000259
NM_001142495
|
MYO5A
|
myosin VA (heavy chain 12, myoxin)
|
|
chr2_-_85788644
|
0.012
|
NM_000821
NM_001142269
|
GGCX
|
gamma-glutamyl carboxylase
|
|
chr2_+_210444364
|
0.012
|
NM_002374
NM_031845
NM_031847
|
MAP2
|
microtubule-associated protein 2
|
|
chr7_+_100797685
|
0.011
|
NM_001283
|
AP1S1
|
adaptor-related protein complex 1, sigma 1 subunit
|
|
chr4_+_6784453
|
0.011
|
NM_001100590
NM_014743
|
KIAA0232
|
KIAA0232
|
|
chr15_-_26874236
|
0.011
|
NM_001191321
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3
|
|
chr4_+_113066552
|
0.011
|
NM_152400
|
C4orf32
|
chromosome 4 open reading frame 32
|
|
chr1_-_247094659
|
0.010
|
NM_015446
|
AHCTF1
AHCTF1P1
|
AT hook containing transcription factor 1
AT hook containing transcription factor 1 pseudogene 1
|
|
chr8_-_139509064
|
0.010
|
NM_015912
|
FAM135B
|
family with sequence similarity 135, member B
|
|
chr2_+_163200582
|
0.010
|
NM_012198
|
GCA
|
grancalcin, EF-hand calcium binding protein
|
|
chr3_-_196695608
|
0.010
|
NM_025163
|
PIGZ
|
phosphatidylinositol glycan anchor biosynthesis, class Z
|
|
chr15_-_56535475
|
0.009
|
NM_022841
|
RFX7
|
regulatory factor X, 7
|
|
chr7_+_4721722
|
0.009
|
NM_001037165
|
FOXK1
|
forkhead box K1
|
|
chr4_+_56212381
|
0.009
|
NM_024592
|
SRD5A3
|
steroid 5 alpha-reductase 3
|
|
chr14_-_20881577
|
0.008
|
NM_007110
|
TEP1
|
telomerase-associated protein 1
|
|
chr16_-_67753186
|
0.008
|
NM_001243650
NM_030819
|
GFOD2
|
glucose-fructose oxidoreductase domain containing 2
|
|
chrX_-_110038992
|
0.008
|
NM_145234
|
CHRDL1
|
chordin-like 1
|
|
chr13_+_50202434
|
0.007
|
NM_138450
|
ARL11
|
ADP-ribosylation factor-like 11
|
|
chr15_+_51633712
|
0.007
|
NM_181789
|
GLDN
|
gliomedin
|
|
chr20_+_48429249
|
0.007
|
NM_015266
|
SLC9A8
|
solute carrier family 9 (sodium/hydrogen exchanger), member 8
|
|
chr12_+_38710556
|
0.006
|
NM_001013620
|
ALG10B
|
asparagine-linked glycosylation 10, alpha-1,2-glucosyltransferase homolog B (yeast)
|
|
chr6_-_134373618
|
0.006
|
NM_145176
|
SLC2A12
|
solute carrier family 2 (facilitated glucose transporter), member 12
|
|
chr6_+_150690027
|
0.006
|
NM_001164694
NM_001164695
NM_203395
|
IYD
|
iodotyrosine deiodinase
|
|
chr4_+_1902352
|
0.006
|
NM_133334
|
WHSC1
|
Wolf-Hirschhorn syndrome candidate 1
|
|
chr2_-_97509733
|
0.006
|
NM_144994
|
ANKRD23
|
ankyrin repeat domain 23
|
|
chr17_-_1303548
|
0.005
|
NM_006761
|
YWHAE
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide
|
|
chr1_+_158149736
|
0.005
|
NM_001766
|
CD1D
|
CD1d molecule
|
|
chr1_-_201465700
|
0.005
|
NM_001193572
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr7_+_138482696
|
0.005
|
NM_001085429
|
TMEM213
|
transmembrane protein 213
|
|
chr5_+_178322915
|
0.005
|
NM_030613
|
ZFP2
|
zinc finger protein 2 homolog (mouse)
|
|
chr1_-_224622000
|
0.005
|
NM_001115113
NM_025160
|
WDR26
|
WD repeat domain 26
|
|
chr13_+_53191601
|
0.004
|
NM_001011724
NM_001011725
|
HNRNPA1L2
|
heterogeneous nuclear ribonucleoprotein A1-like 2
|
|
chr6_+_7590405
|
0.004
|
NM_152551
|
SNRNP48
|
small nuclear ribonucleoprotein 48kDa (U11/U12)
|
|
chr3_+_42947401
|
0.004
|
NM_207404
|
ZNF662
|
zinc finger protein 662
|
|
chr1_+_231298673
|
0.004
|
NM_001004342
|
TRIM67
|
tripartite motif containing 67
|
|
chr1_-_8086392
|
0.004
|
NM_018948
|
ERRFI1
|
ERBB receptor feedback inhibitor 1
|
|
chr7_-_51384458
|
0.004
|
NM_015198
|
COBL
|
cordon-bleu homolog (mouse)
|
|
chr20_+_43104513
|
0.004
|
NM_001039199
NM_024331
|
TTPAL
|
tocopherol (alpha) transfer protein-like
|
|
chr3_+_143692166
|
0.004
|
NM_001134470
|
C3orf58
|
chromosome 3 open reading frame 58
|
|
chr1_-_173572179
|
0.003
|
NM_178527
|
SLC9A11
|
solute carrier family 9, member 11
|
|
chr3_+_42947649
|
0.003
|
NM_001134656
|
ZNF662
|
zinc finger protein 662
|
|
chr1_-_201476248
|
0.003
|
NM_001193570
NM_004078
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr7_+_23286277
|
0.003
|
NM_001005340
NM_002510
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr1_-_201475966
|
0.003
|
NM_001193571
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr1_-_230513390
|
0.003
|
NM_024554
|
PGBD5
|
piggyBac transposable element derived 5
|
|
chr14_-_94942513
|
0.003
|
NM_001042518
NM_175739
|
SERPINA9
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 9
|
|
chr3_-_87040246
|
0.002
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr17_+_5390212
|
0.002
|
NM_024039
|
MIS12
|
MIS12, MIND kinetochore complex component, homolog (S. pombe)
|
|
chr3_-_45017600
|
0.002
|
NM_001135179
NM_016598
|
ZDHHC3
|
zinc finger, DHHC-type containing 3
|
|
chr13_-_46425756
|
0.001
|
NM_198849
|
SIAH3
|
seven in absentia homolog 3 (Drosophila)
|
|
chr10_-_53459309
|
0.001
|
NM_015235
|
CSTF2T
|
cleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa, tau variant
|
|
chr20_-_57617836
|
0.001
|
NM_016045
|
SLMO2
|
slowmo homolog 2 (Drosophila)
|
|
chr11_+_18720342
|
0.000
|
NM_153347
|
TMEM86A
|
transmembrane protein 86A
|
|
chr6_+_76458888
|
0.000
|
NM_004999
|
MYO6
|
myosin VI
|