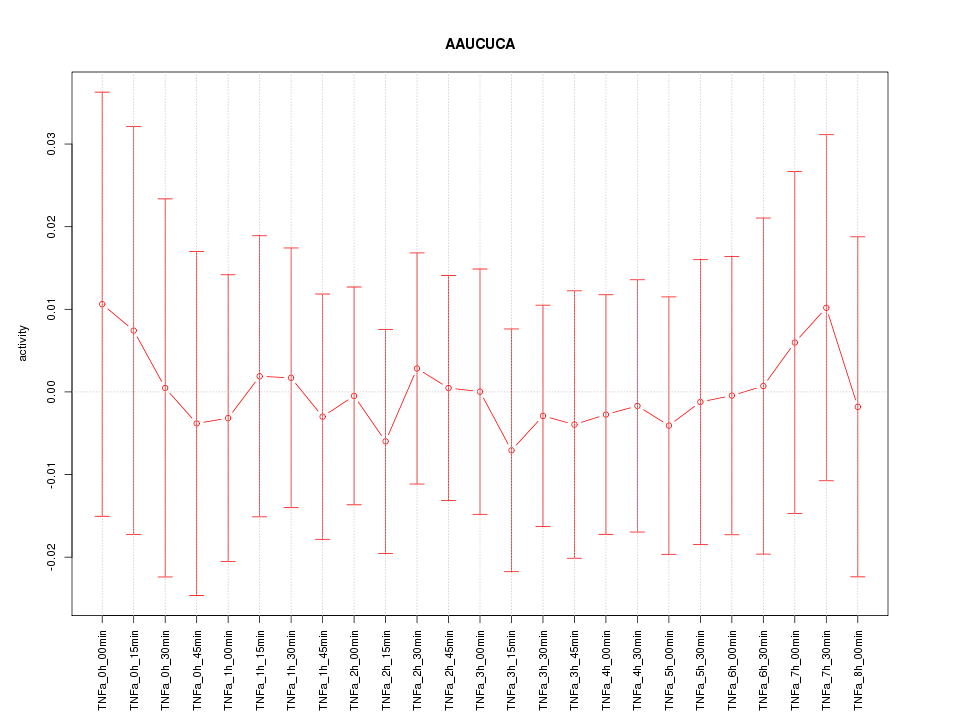
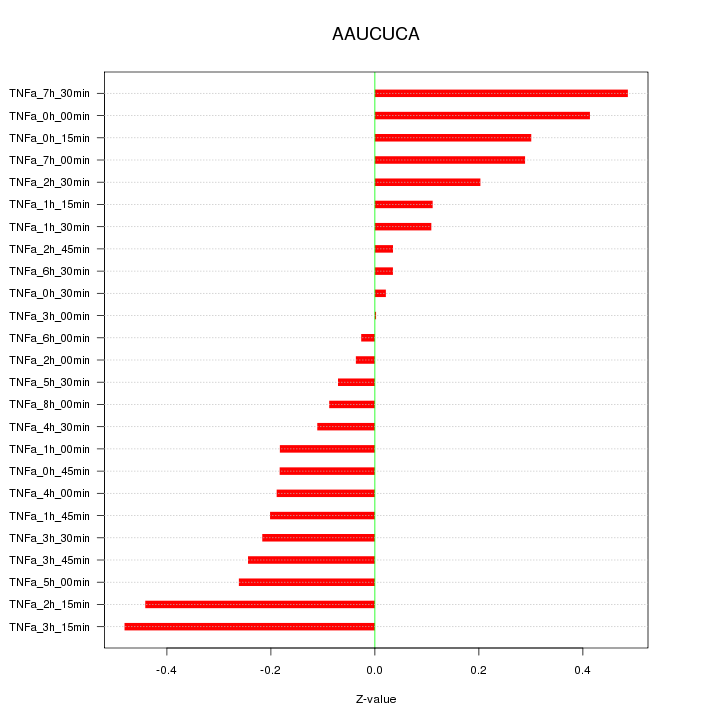
|
chr3_+_170075449
|
0.137
|
NM_001145098
NM_005414
|
SKIL
|
SKI-like oncogene
|
|
chr12_-_95044205
|
0.091
|
NM_020698
|
TMCC3
|
transmembrane and coiled-coil domain family 3
|
|
chrX_-_20284708
|
0.091
|
NM_004586
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3
|
|
chr4_+_89299874
|
0.090
|
NM_001165136
NM_017912
|
HERC6
|
hect domain and RLD 6
|
|
chr6_+_147829784
|
0.090
|
NM_001030060
|
SAMD5
|
sterile alpha motif domain containing 5
|
|
chr1_-_53793743
|
0.085
|
NM_001018054
NM_004631
NM_017522
NM_033300
|
LRP8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor
|
|
chr10_-_97321094
|
0.080
|
NM_015385
NM_024991
|
SORBS1
|
sorbin and SH3 domain containing 1
|
|
chr18_-_22932109
|
0.080
|
NM_015461
|
ZNF521
|
zinc finger protein 521
|
|
chr3_+_170077410
|
0.077
|
NM_001145097
NM_001248008
|
SKIL
|
SKI-like oncogene
|
|
chr1_-_151882360
|
0.076
|
NM_053055
|
THEM4
|
thioesterase superfamily member 4
|
|
chr6_+_110501591
|
0.075
|
NM_015891
|
CDC40
|
cell division cycle 40 homolog (S. cerevisiae)
|
|
chr2_-_151344145
|
0.074
|
NM_005168
|
RND3
|
Rho family GTPase 3
|
|
chr11_+_111848008
|
0.073
|
NM_033425
|
DIXDC1
|
DIX domain containing 1
|
|
chr12_-_50236868
|
0.070
|
NM_181708
|
BCDIN3D
|
BCDIN3 domain containing
|
|
chr4_+_175204827
|
0.070
|
NM_001040157
|
CEP44
|
centrosomal protein 44kDa
|
|
chr9_+_4662285
|
0.070
|
NM_203453
|
PPAPDC2
|
phosphatidic acid phosphatase type 2 domain containing 2
|
|
chr3_-_125094003
|
0.069
|
NM_021964
|
ZNF148
|
zinc finger protein 148
|
|
chr7_+_12250818
|
0.068
|
NM_001134232
NM_018374
|
TMEM106B
|
transmembrane protein 106B
|
|
chr5_+_149151502
|
0.066
|
NM_001172699
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr5_+_149109814
|
0.065
|
NM_001172698
NM_133263
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr10_-_97200895
|
0.063
|
NM_001034954
NM_001034955
NM_001034956
NM_001034957
NM_006434
|
SORBS1
|
sorbin and SH3 domain containing 1
|
|
chr18_+_59854514
|
0.062
|
NM_020854
|
KIAA1468
|
KIAA1468
|
|
chr6_+_42788793
|
0.061
|
NM_015349
|
KIAA0240
|
KIAA0240
|
|
chr6_-_138428477
|
0.059
|
NM_022121
|
PERP
|
PERP, TP53 apoptosis effector
|
|
chr3_+_45927995
|
0.058
|
NM_006641
NM_031200
|
CCR9
|
chemokine (C-C motif) receptor 9
|
|
chr7_-_105319608
|
0.057
|
NM_138495
|
ATXN7L1
|
ataxin 7-like 1
|
|
chr1_-_220263150
|
0.057
|
NM_006085
|
BPNT1
|
3'(2'), 5'-bisphosphate nucleotidase 1
|
|
chr2_+_148778573
|
0.056
|
NM_018328
|
MBD5
|
methyl-CpG binding domain protein 5
|
|
chr7_-_91509979
|
0.056
|
NM_006980
|
MTERF
|
mitochondrial transcription termination factor
|
|
chr10_-_95462311
|
0.055
|
NM_145246
|
FRA10AC1
|
fragile site, folic acid type, rare, fra(10)(q23.3) or fra(10)(q24.2) candidate 1
|
|
chr1_-_205782135
|
0.055
|
NM_173854
|
SLC41A1
|
solute carrier family 41, member 1
|
|
chr5_+_130506635
|
0.050
|
NM_181705
|
LYRM7
|
Lyrm7 homolog (mouse)
|
|
chr7_+_35840595
|
0.050
|
NM_001011553
NM_001242956
NM_001788
|
SEPT7
|
septin 7
|
|
chr13_-_21476895
|
0.049
|
NM_022459
|
XPO4
|
exportin 4
|
|
chr3_-_160117314
|
0.048
|
NM_001190241
NM_020800
|
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas)
|
|
chr4_+_99916787
|
0.047
|
NM_015143
|
METAP1
|
methionyl aminopeptidase 1
|
|
chr4_-_88141614
|
0.047
|
NM_020803
|
KLHL8
|
kelch-like 8 (Drosophila)
|
|
chr5_+_139493689
|
0.046
|
NM_005859
|
PURA
|
purine-rich element binding protein A
|
|
chr3_-_160101443
|
0.045
|
NM_001190242
|
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas)
|
|
chr2_+_109204745
|
0.044
|
NM_004987
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr2_+_109204904
|
0.044
|
NM_001193488
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr15_+_91643429
|
0.044
|
NM_014848
|
SV2B
|
synaptic vesicle glycoprotein 2B
|
|
chr8_-_87520981
|
0.043
|
NM_016033
|
FAM82B
|
family with sequence similarity 82, member B
|
|
chr3_+_108308336
|
0.043
|
NM_014648
|
DZIP3
|
DAZ interacting protein 3, zinc finger
|
|
chr12_+_123237302
|
0.042
|
NM_003677
|
DENR
|
density-regulated protein
|
|
chrX_-_109683457
|
0.042
|
NM_001171689
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr6_+_7590405
|
0.042
|
NM_152551
|
SNRNP48
|
small nuclear ribonucleoprotein 48kDa (U11/U12)
|
|
chrX_-_109561314
|
0.042
|
NM_001025580
NM_015365
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr8_-_110342107
|
0.041
|
NM_001128211
|
NUDCD1
|
NudC domain containing 1
|
|
chr15_-_43662107
|
0.041
|
NM_152455
|
ZSCAN29
|
zinc finger and SCAN domain containing 29
|
|
chr3_+_39851028
|
0.041
|
NM_015460
|
MYRIP
|
myosin VIIA and Rab interacting protein
|
|
chr11_+_74459900
|
0.041
|
NM_001098638
|
RNF169
|
ring finger protein 169
|
|
chr8_-_110346272
|
0.041
|
NM_032869
|
NUDCD1
|
NudC domain containing 1
|
|
chr6_-_82462374
|
0.040
|
NM_017633
|
FAM46A
|
family with sequence similarity 46, member A
|
|
chr2_+_109237679
|
0.040
|
NM_001193484
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr12_-_31743822
|
0.039
|
NM_144973
|
DENND5B
|
DENN/MADD domain containing 5B
|
|
chr2_+_109150807
|
0.039
|
NM_001193483
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr16_-_57520326
|
0.039
|
NM_018110
|
DOK4
|
docking protein 4
|
|
chr7_-_115670828
|
0.039
|
NM_001018058
NM_012252
|
TFEC
|
transcription factor EC
|
|
chr19_+_54024176
|
0.039
|
NM_018555
|
ZNF331
|
zinc finger protein 331
|
|
chr2_+_109271267
|
0.039
|
NM_001193485
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr2_+_178257371
|
0.039
|
NM_003659
|
AGPS
|
alkylglycerone phosphate synthase
|
|
chrX_+_119738542
|
0.038
|
NM_001137554
|
MCTS1
|
malignant T cell amplified sequence 1
|
|
chr1_+_236687026
|
0.037
|
NM_201544
|
LGALS8
|
lectin, galactoside-binding, soluble, 8
|
|
chr11_+_111807857
|
0.037
|
NM_001037954
|
DIXDC1
|
DIX domain containing 1
|
|
chr12_-_89919182
|
0.037
|
NM_001199777
|
POC1B
|
POC1 centriolar protein homolog B (Chlamydomonas)
|
|
chr13_-_31040035
|
0.036
|
NM_002128
|
HMGB1
|
high mobility group box 1
|
|
chr4_+_81952118
|
0.035
|
NM_001201
|
BMP3
|
bone morphogenetic protein 3
|
|
chr17_-_7382943
|
0.035
|
NM_001128833
|
ZBTB4
|
zinc finger and BTB domain containing 4
|
|
chr3_-_121553907
|
0.034
|
NM_001023570
NM_001023571
|
IQCB1
|
IQ motif containing B1
|
|
chr12_-_10022457
|
0.034
|
NM_005127
|
CLEC2B
|
C-type lectin domain family 2, member B
|
|
chr19_+_54041332
|
0.034
|
NM_001079906
NM_001253798
NM_001253799
|
ZNF331
|
zinc finger protein 331
|
|
chr8_+_1922030
|
0.034
|
NM_014867
|
KBTBD11
|
kelch repeat and BTB (POZ) domain containing 11
|
|
chr5_-_137549029
|
0.034
|
NM_004661
|
CDC23
|
cell division cycle 23 homolog (S. cerevisiae)
|
|
chrX_+_119737743
|
0.034
|
NM_014060
|
MCTS1
|
malignant T cell amplified sequence 1
|
|
chr2_+_109223600
|
0.033
|
NM_001193482
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr10_+_11206928
|
0.033
|
NM_001025076
NM_001083591
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr1_+_236686738
|
0.032
|
NM_006499
NM_201543
|
LGALS8
|
lectin, galactoside-binding, soluble, 8
|
|
chr3_+_101498028
|
0.032
|
NM_145037
|
FAM55C
|
family with sequence similarity 55, member C
|
|
chr1_+_236681513
|
0.032
|
NM_201545
|
LGALS8
|
lectin, galactoside-binding, soluble, 8
|
|
chr20_+_54933982
|
0.032
|
NM_080821
|
FAM210B
|
family with sequence similarity 210, member B
|
|
chr17_+_64298844
|
0.031
|
NM_002737
|
PRKCA
|
protein kinase C, alpha
|
|
chr15_+_65914269
|
0.031
|
NM_004727
|
SLC24A1
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 1
|
|
chr17_-_47755340
|
0.030
|
NM_001007226
NM_001007227
NM_001007228
NM_001007229
NM_001007230
NM_003563
|
SPOP
|
speckle-type POZ protein
|
|
chr17_+_59477169
|
0.030
|
NM_005994
|
TBX2
|
T-box 2
|
|
chr2_+_175260455
|
0.030
|
NM_001193528
NM_024583
|
SCRN3
|
secernin 3
|
|
chr1_+_24882565
|
0.029
|
NM_001010980
|
C1orf130
|
chromosome 1 open reading frame 130
|
|
chr9_-_14693459
|
0.029
|
NM_178566
|
ZDHHC21
|
zinc finger, DHHC-type containing 21
|
|
chr17_+_53342320
|
0.029
|
NM_002126
|
HLF
|
hepatic leukemia factor
|
|
chr4_-_121993568
|
0.029
|
NM_024574
|
NDNF
|
neuron-derived neurotrophic factor
|
|
chr6_-_134373618
|
0.029
|
NM_145176
|
SLC2A12
|
solute carrier family 2 (facilitated glucose transporter), member 12
|
|
chr3_+_142342265
|
0.029
|
NM_002670
|
PLS1
|
plastin 1
|
|
chr19_+_52901101
|
0.028
|
NM_032423
|
ZNF528
|
zinc finger protein 528
|
|
chr16_+_8768403
|
0.028
|
NM_020686
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr8_+_67341244
|
0.028
|
NM_015169
|
RRS1
|
RRS1 ribosome biogenesis regulator homolog (S. cerevisiae)
|
|
chr1_-_67896098
|
0.027
|
NM_001018067
NM_001018068
NM_001018069
NM_015640
|
SERBP1
|
SERPINE1 mRNA binding protein 1
|
|
chr11_-_64764388
|
0.027
|
NM_138456
|
BATF2
|
basic leucine zipper transcription factor, ATF-like 2
|
|
chr5_+_68788387
|
0.027
|
NM_001205254
|
OCLN
|
occludin
|
|
chr20_+_18122867
|
0.027
|
NM_020536
|
CSRP2BP
|
CSRP2 binding protein
|
|
chr1_-_155532322
|
0.027
|
NM_018489
|
ASH1L
|
ash1 (absent, small, or homeotic)-like (Drosophila)
|
|
chr2_+_139259349
|
0.027
|
NM_001001664
|
SPOPL
|
speckle-type POZ protein-like
|
|
chr17_+_48046561
|
0.027
|
NM_138281
|
DLX4
|
distal-less homeobox 4
|
|
chr8_-_146126776
|
0.027
|
NM_001109689
NM_021061
|
ZNF250
|
zinc finger protein 250
|
|
chr2_+_85980600
|
0.027
|
NM_032827
|
ATOH8
|
atonal homolog 8 (Drosophila)
|
|
chr20_+_43514340
|
0.026
|
NM_003404
NM_139323
|
YWHAB
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide
|
|
chr18_-_61034350
|
0.026
|
NM_002035
|
KDSR
|
3-ketodihydrosphingosine reductase
|
|
chr3_+_142375738
|
0.026
|
NM_001172312
|
PLS1
|
plastin 1
|
|
chr1_-_52456292
|
0.026
|
NM_002867
|
RAB3B
|
RAB3B, member RAS oncogene family
|
|
chr5_-_58334936
|
0.026
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr16_+_8814567
|
0.026
|
NM_001127448
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr4_-_85887543
|
0.026
|
NM_014991
|
WDFY3
|
WD repeat and FYVE domain containing 3
|
|
chr14_+_102228134
|
0.025
|
NM_001161725
NM_001161726
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr10_+_11047253
|
0.025
|
NM_001025077
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr6_-_80247103
|
0.025
|
NM_001122769
NM_181714
|
LCA5
|
Leber congenital amaurosis 5
|
|
chr10_+_46222647
|
0.025
|
NM_001169106
NM_001169107
NM_015262
|
FAM21C
|
family with sequence similarity 21, member C
|
|
chr10_+_11059854
|
0.025
|
NM_006561
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr11_-_132812870
|
0.025
|
NM_002545
|
OPCML
|
opioid binding protein/cell adhesion molecule-like
|
|
chr5_-_59189620
|
0.024
|
NM_001104631
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr19_+_54058493
|
0.024
|
NM_001079907
|
ZNF331
|
zinc finger protein 331
|
|
chr8_-_144923019
|
0.024
|
NM_178564
|
NRBP2
|
nuclear receptor binding protein 2
|
|
chr16_+_8806825
|
0.024
|
NM_000663
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr2_+_95831176
|
0.024
|
NM_001017396
NM_021088
|
ZNF2
|
zinc finger protein 2
|
|
chr10_+_51827654
|
0.024
|
NM_001005751
|
FAM21C
FAM21A
|
family with sequence similarity 21, member C
family with sequence similarity 21, member A
|
|
chr3_+_142315205
|
0.024
|
NM_001145319
|
PLS1
|
plastin 1
|
|
chr10_-_104473957
|
0.024
|
NM_004311
|
ARL3
|
ADP-ribosylation factor-like 3
|
|
chr1_-_89738477
|
0.024
|
NM_001134486
NM_052942
|
GBP5
|
guanylate binding protein 5
|
|
chr5_-_59783885
|
0.024
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_-_58295758
|
0.023
|
NM_001197223
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr11_+_108093558
|
0.023
|
NM_000051
|
ATM
|
ataxia telangiectasia mutated
|
|
chr3_+_101498285
|
0.023
|
NM_001134456
|
FAM55C
|
family with sequence similarity 55, member C
|
|
chr22_+_21213248
|
0.023
|
NM_004782
|
SNAP29
|
synaptosomal-associated protein, 29kDa
|
|
chr9_-_15510209
|
0.023
|
NM_021144
|
PSIP1
|
PC4 and SFRS1 interacting protein 1
|
|
chr3_-_121264780
|
0.023
|
NM_199420
|
POLQ
|
polymerase (DNA directed), theta
|
|
chr5_-_58652671
|
0.023
|
NM_001197219
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_-_58571916
|
0.022
|
NM_001197220
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr11_+_111944967
|
0.022
|
NM_001082969
NM_001082970
NM_018195
|
C11orf57
|
chromosome 11 open reading frame 57
|
|
chr2_-_203776948
|
0.022
|
NM_018256
|
WDR12
|
WD repeat domain 12
|
|
chr15_-_33360084
|
0.022
|
NM_001103184
|
FMN1
|
formin 1
|
|
chr6_+_30951484
|
0.021
|
NM_001010909
|
MUC21
|
mucin 21, cell surface associated
|
|
chr4_-_113437327
|
0.021
|
NM_024019
|
NEUROG2
|
neurogenin 2
|
|
chr16_-_19729417
|
0.021
|
NM_001012991
|
C16orf88
|
chromosome 16 open reading frame 88
|
|
chr11_+_96123120
|
0.021
|
NM_003772
|
JRKL
|
jerky homolog-like (mouse)
|
|
chr11_-_130184459
|
0.021
|
NM_014155
|
ZBTB44
|
zinc finger and BTB domain containing 44
|
|
chr5_+_122424840
|
0.021
|
NM_001136239
|
PRDM6
|
PR domain containing 6
|
|
chr5_+_68800003
|
0.021
|
NM_001205255
|
OCLN
|
occludin
|
|
chr1_-_109506016
|
0.021
|
NM_001048210
NM_015127
|
CLCC1
AKNAD1
|
chloride channel CLIC-like 1
AKNA domain containing 1
|
|
chr17_+_37793332
|
0.021
|
NM_001165937
NM_001165938
NM_006804
|
STARD3
|
StAR-related lipid transfer (START) domain containing 3
|
|
chr12_+_106994914
|
0.020
|
NM_001206691
|
RFX4
|
regulatory factor X, 4 (influences HLA class II expression)
|
|
chr17_-_7387513
|
0.020
|
NM_020899
|
ZBTB4
|
zinc finger and BTB domain containing 4
|
|
chr17_+_46018888
|
0.020
|
NM_018129
|
PNPO
|
pyridoxamine 5'-phosphate oxidase
|
|
chr1_-_236030219
|
0.019
|
NM_000081
|
LYST
|
lysosomal trafficking regulator
|
|
chr16_+_69599868
|
0.019
|
NM_001113178
NM_006599
NM_138713
NM_138714
NM_173214
NM_173215
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr2_+_135676387
|
0.019
|
NM_001241
NM_058241
|
CCNT2
|
cyclin T2
|
|
chr3_+_196295708
|
0.019
|
NM_001105573
|
FBXO45
|
F-box protein 45
|
|
chr22_-_46933066
|
0.019
|
NM_014246
|
CELSR1
|
cadherin, EGF LAG seven-pass G-type receptor 1 (flamingo homolog, Drosophila)
|
|
chr9_+_125133228
|
0.018
|
NM_000962
NM_080591
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr17_-_26941178
|
0.018
|
NM_001174103
|
SGK494
|
uncharacterized serine/threonine-protein kinase SgK494
|
|
chr11_-_69490093
|
0.018
|
NM_153451
|
ORAOV1
|
oral cancer overexpressed 1
|
|
chrX_-_7895424
|
0.017
|
NM_004650
|
PNPLA4
|
patatin-like phospholipase domain containing 4
|
|
chr17_-_16118789
|
0.017
|
NM_006311
|
NCOR1
|
nuclear receptor corepressor 1
|
|
chr3_-_69129502
|
0.017
|
NM_003968
NM_198195
|
UBA3
|
ubiquitin-like modifier activating enzyme 3
|
|
chr5_-_10761344
|
0.016
|
NM_004394
|
DAP
|
death-associated protein
|
|
chr11_-_133402227
|
0.016
|
NM_001012393
|
OPCML
|
opioid binding protein/cell adhesion molecule-like
|
|
chr1_-_67520028
|
0.016
|
NM_015139
|
SLC35D1
|
solute carrier family 35 (UDP-glucuronic acid/UDP-N-acetylgalactosamine dual transporter), member D1
|
|
chr1_-_144931967
|
0.016
|
NM_001002811
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr2_+_26987122
|
0.016
|
NM_017877
|
CENPA
C2orf18
|
centromere protein A
chromosome 2 open reading frame 18
|
|
chr5_-_59064437
|
0.015
|
NM_001197218
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr6_-_107780682
|
0.015
|
NM_020381
|
PDSS2
|
prenyl (decaprenyl) diphosphate synthase, subunit 2
|
|
chr10_+_112404104
|
0.015
|
NM_001134363
|
RBM20
|
RNA binding motif protein 20
|
|
chrX_-_7895764
|
0.015
|
NM_001142389
NM_001172672
|
PNPLA4
|
patatin-like phospholipase domain containing 4
|
|
chr15_-_83953465
|
0.015
|
NM_001717
|
BNC1
|
basonuclin 1
|
|
chr16_-_10276115
|
0.014
|
NM_001134407
|
GRIN2A
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A
|
|
chr16_-_10276610
|
0.014
|
NM_000833
|
GRIN2A
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A
|
|
chr15_+_91643181
|
0.014
|
NM_001167580
|
SV2B
|
synaptic vesicle glycoprotein 2B
|
|
chr8_-_57233240
|
0.014
|
NM_138969
|
SDR16C5
|
short chain dehydrogenase/reductase family 16C, member 5
|
|
chr9_+_115513050
|
0.014
|
NM_001012994
|
SNX30
|
sorting nexin family member 30
|
|
chr13_-_100624162
|
0.013
|
NM_033132
|
ZIC5
|
Zic family member 5
|
|
chr15_+_63481713
|
0.013
|
NM_016530
|
RAB8B
|
RAB8B, member RAS oncogene family
|
|
chr4_+_145567147
|
0.013
|
NM_022475
|
HHIP
|
hedgehog interacting protein
|
|
chr2_+_113885137
|
0.013
|
NM_173842
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr5_-_34043234
|
0.013
|
NM_030945
NM_181435
|
C1QTNF3
|
C1q and tumor necrosis factor related protein 3
|
|
chr5_-_58882274
|
0.013
|
NM_006203
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr3_+_40566341
|
0.013
|
NM_001098414
NM_198484
|
ZNF621
|
zinc finger protein 621
|
|
chr14_-_35183722
|
0.012
|
NM_001243645
NM_138638
|
CFL2
|
cofilin 2 (muscle)
|
|
chr15_-_42840992
|
0.012
|
NM_153260
|
LRRC57
|
leucine rich repeat containing 57
|
|
chr3_+_141043054
|
0.012
|
NM_001080412
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr15_+_22833394
|
0.012
|
NM_001102610
NM_052903
|
TUBGCP5
|
tubulin, gamma complex associated protein 5
|
|
chr20_+_11898536
|
0.012
|
NM_014962
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr4_+_76649826
|
0.012
|
NM_003715
|
USO1
|
USO1 vesicle docking protein homolog (yeast)
|
|
chr12_+_27863705
|
0.012
|
NM_001190864
NM_021821
|
MRPS35
|
mitochondrial ribosomal protein S35
|
|
chr3_-_142166782
|
0.012
|
NM_001042604
NM_019001
|
XRN1
|
5'-3' exoribonuclease 1
|
|
chr3_+_42642105
|
0.012
|
NM_005385
|
NKTR
|
natural killer-tumor recognition sequence
|
|
chr16_-_4292052
|
0.011
|
NM_001098814
|
SRL
|
sarcalumenin
|
|
chr12_-_57030083
|
0.011
|
NM_013449
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A
|
|
chr17_-_29641094
|
0.011
|
NM_006495
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr17_-_73975507
|
0.011
|
NM_001185039
NM_004035
NM_007292
|
ACOX1
|
acyl-CoA oxidase 1, palmitoyl
|
|
chr5_+_68788118
|
0.010
|
NM_002538
|
OCLN
|
occludin
|
|
chr6_-_24666818
|
0.010
|
NM_016614
|
TDP2
|
tyrosyl-DNA phosphodiesterase 2
|
|
chr1_-_68915631
|
0.010
|
NM_000329
|
RPE65
|
retinal pigment epithelium-specific protein 65kDa
|
|
chr1_+_78956727
|
0.010
|
NM_000959
NM_001039585
|
PTGFR
|
prostaglandin F receptor (FP)
|