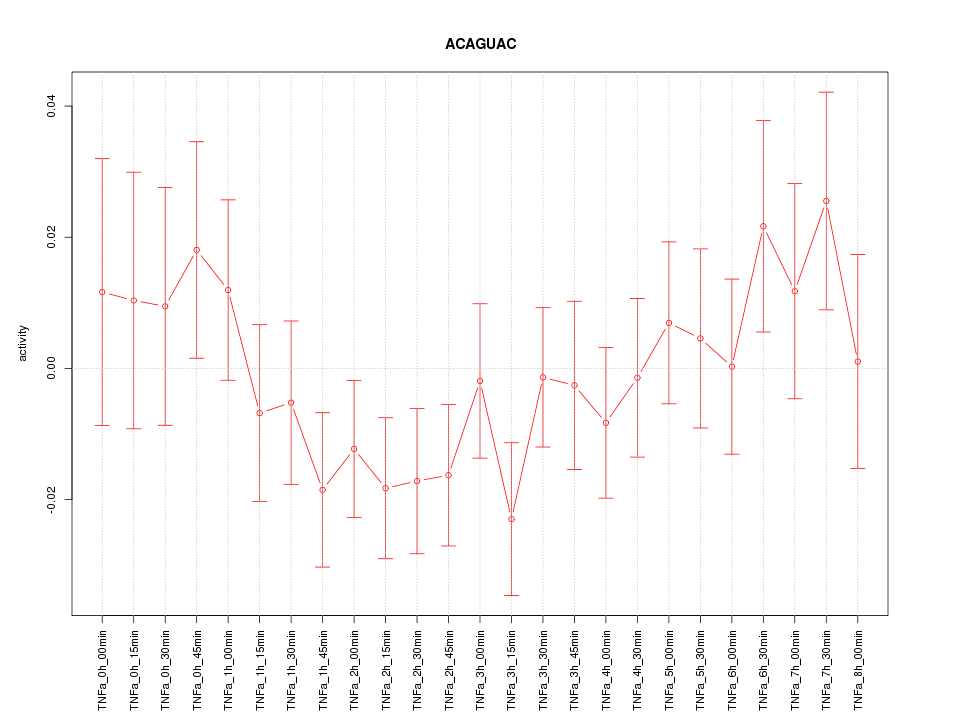
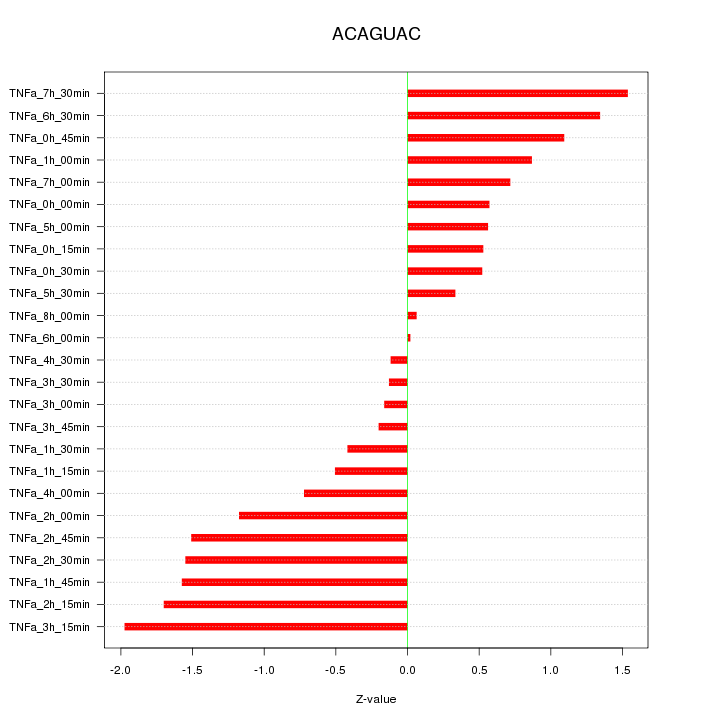
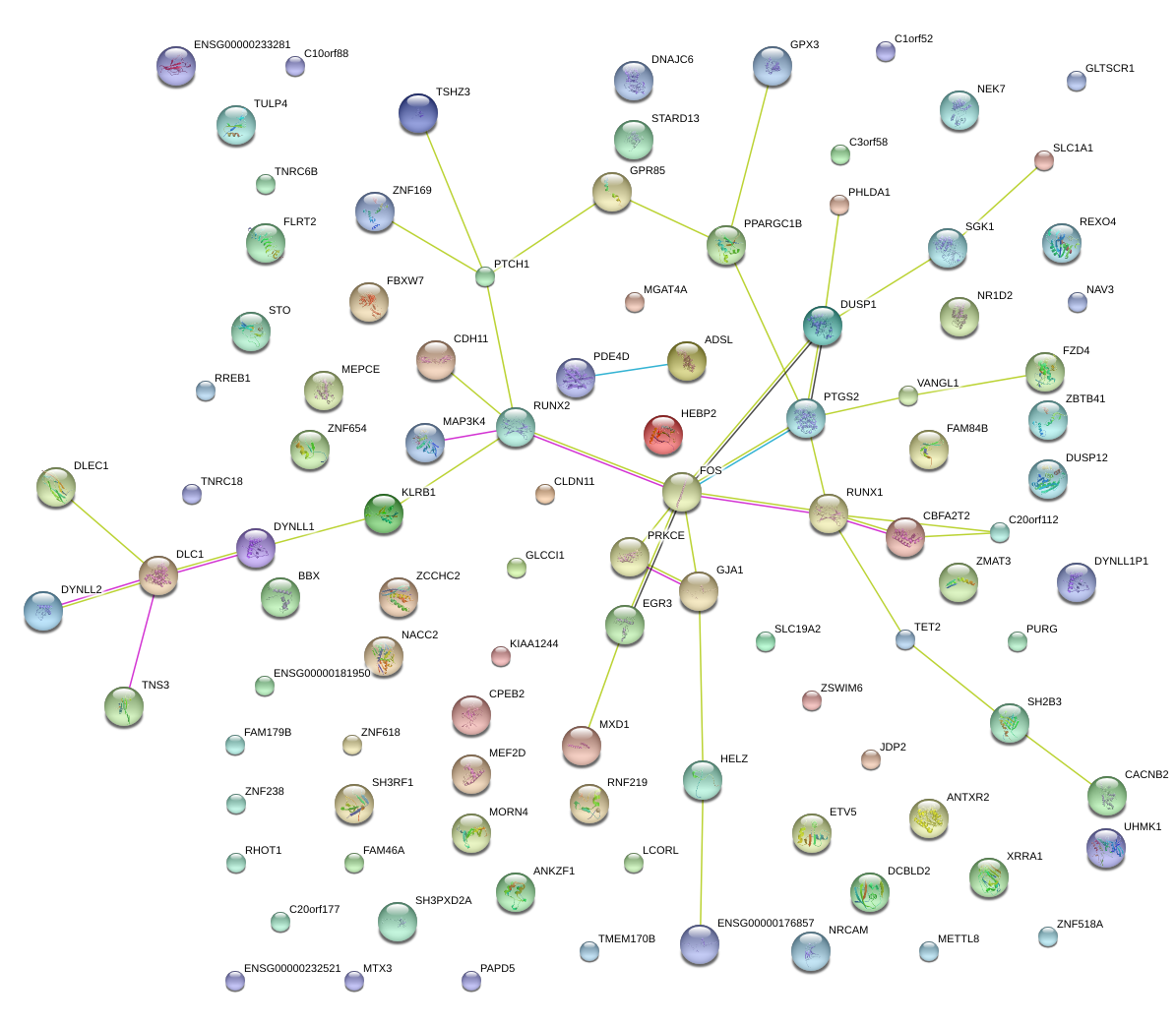
|
chr20_+_58515408
|
2.087
|
NM_001190827
NM_022106
|
C20orf177
|
chromosome 20 open reading frame 177
|
|
chr20_+_58508818
|
2.061
|
NM_001190826
|
C20orf177
|
chromosome 20 open reading frame 177
|
|
chr1_-_186649421
|
2.024
|
NM_000963
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr6_-_82462374
|
1.488
|
NM_017633
|
FAM46A
|
family with sequence similarity 46, member A
|
|
chr6_-_134497069
|
1.434
|
NM_001143678
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr6_-_134496006
|
1.420
|
NM_005627
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr6_-_134498973
|
1.411
|
NM_001143677
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr3_+_88188261
|
1.401
|
NM_018293
|
ZNF654
|
zinc finger protein 654
|
|
chr6_-_134639195
|
1.384
|
NM_001143676
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr3_-_185826748
|
1.313
|
NM_004454
|
ETV5
|
ets variant 5
|
|
chr5_-_59189620
|
1.305
|
NM_001104631
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr1_-_169455099
|
1.249
|
NM_006996
|
SLC19A2
|
solute carrier family 19 (thiamine transporter), member 2
|
|
chr5_-_58334936
|
1.246
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr8_-_22550057
|
1.231
|
NM_001199881
|
EGR3
|
early growth response 3
|
|
chr8_-_22549901
|
1.225
|
NM_001199880
|
EGR3
|
early growth response 3
|
|
chr11_-_86666211
|
1.212
|
NM_012193
|
FZD4
|
frizzled family receptor 4
|
|
chr2_+_45878912
|
1.208
|
NM_005400
|
PRKCE
|
protein kinase C, epsilon
|
|
chr8_-_22550708
|
1.174
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr3_-_178789597
|
1.171
|
NM_022470
NM_152240
|
ZMAT3
|
zinc finger, matrin-type 3
|
|
chr5_-_58571916
|
1.152
|
NM_001197220
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_-_58652671
|
1.142
|
NM_001197219
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_-_58295758
|
1.129
|
NM_001197223
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr16_-_65155881
|
1.126
|
NM_001797
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr5_-_59783885
|
1.118
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_-_58882274
|
1.065
|
NM_006203
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_-_59064437
|
1.033
|
NM_001197218
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_-_172198160
|
1.023
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chr5_+_176560056
|
1.015
|
NM_172349
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr13_-_33760215
|
0.998
|
NM_001243466
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr13_-_33859835
|
0.979
|
NM_178006
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr13_-_33780142
|
0.976
|
NM_001243474
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr5_+_60628085
|
0.962
|
NM_020928
|
ZSWIM6
|
zinc finger, SWIM-type containing 6
|
|
chr20_-_31071187
|
0.894
|
NM_080616
|
C20orf112
|
chromosome 20 open reading frame 112
|
|
chr19_-_31840118
|
0.874
|
NM_020856
|
TSHZ3
|
teashirt zinc finger homeobox 3
|
|
chr10_+_97889467
|
0.832
|
NM_014803
|
ZNF518A
|
zinc finger protein 518A
|
|
chr5_+_176560627
|
0.823
|
NM_022455
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr18_+_60190657
|
0.747
|
NM_017742
|
ZCCHC2
|
zinc finger, CCHC domain containing 2
|
|
chr14_+_75745480
|
0.717
|
NM_005252
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog
|
|
chr1_-_197169635
|
0.713
|
NM_194314
|
ZBTB41
|
zinc finger and BTB domain containing 41
|
|
chr14_+_85996454
|
0.666
|
NM_013231
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr7_-_108096693
|
0.653
|
NM_001193582
NM_001193583
NM_001193584
NM_005010
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr2_-_172291227
|
0.646
|
NM_024770
|
METTL8
|
methyltransferase like 8
|
|
chr22_+_40440664
|
0.645
|
NM_001024843
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr4_+_106067754
|
0.636
|
NM_001127208
|
TET2
|
tet methylcytosine dioxygenase 2
|
|
chr2_+_220094478
|
0.621
|
NM_018089
NM_001042410
|
ANKZF1
|
ankyrin repeat and zinc finger domain containing 1
|
|
chr4_-_18023358
|
0.597
|
NM_001166139
NM_153686
|
LCORL
|
ligand dependent nuclear receptor corepressor-like
|
|
chr7_-_5463176
|
0.592
|
NM_001080495
|
TNRC18
|
trinucleotide repeat containing 18
|
|
chr6_+_121756744
|
0.586
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr4_-_153273683
|
0.584
|
NM_018315
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr4_-_153456152
|
0.583
|
NM_033632
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr7_-_47621741
|
0.578
|
NM_022748
|
TNS3
|
tensin 3
|
|
chr6_+_7107986
|
0.569
|
NM_001003698
NM_001003699
NM_001003700
|
RREB1
|
ras responsive element binding protein 1
|
|
chr5_+_149151502
|
0.556
|
NM_001172699
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr14_+_75894919
|
0.546
|
NM_001135048
NM_130469
|
JDP2
|
Jun dimerization protein 2
|
|
chr5_+_149109814
|
0.542
|
NM_001172698
NM_133263
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr7_-_112727832
|
0.537
|
NM_001146265
NM_018970
|
GPR85
|
G protein-coupled receptor 85
|
|
chr14_+_75894508
|
0.533
|
NM_001135047
|
JDP2
|
Jun dimerization protein 2
|
|
chr10_+_18549583
|
0.531
|
NM_000724
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr6_+_7107799
|
0.530
|
NM_001168344
|
RREB1
|
ras responsive element binding protein 1
|
|
chr9_-_138987096
|
0.528
|
NM_144653
|
NACC2
|
NACC family member 2, BEN and BTB (POZ) domain containing
|
|
chr8_-_30891230
|
0.527
|
NM_001015508
|
PURG
|
purine-rich element binding protein G
|
|
chr14_+_75898836
|
0.518
|
NM_001135049
|
JDP2
|
Jun dimerization protein 2
|
|
chr16_+_50186809
|
0.516
|
NM_001040284
NM_001040285
|
PAPD5
|
PAP associated domain containing 5
|
|
chr9_+_4490193
|
0.515
|
NM_004170
|
SLC1A1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1
|
|
chr9_-_98269480
|
0.508
|
NM_001083607
|
PTCH1
|
patched 1
|
|
chr4_-_153303663
|
0.505
|
NM_001013415
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr7_-_112726143
|
0.504
|
NM_001146266
NM_001146267
|
GPR85
|
G protein-coupled receptor 85
|
|
chr6_+_158733691
|
0.497
|
NM_001007466
NM_020245
|
TULP4
|
tubby like protein 4
|
|
chr6_+_161412807
|
0.493
|
NM_005922
NM_006724
|
MAP3K4
|
mitogen-activated protein kinase kinase kinase 4
|
|
chr5_-_79286793
|
0.487
|
NM_001010891
NM_001167741
|
MTX3
|
metaxin 3
|
|
chr10_-_99393175
|
0.487
|
NM_178832
|
MORN4
|
MORN repeat containing 4
|
|
chr1_+_244216506
|
0.485
|
NM_006352
|
ZNF238
|
zinc finger protein 238
|
|
chr1_+_244214552
|
0.480
|
NM_205768
|
ZNF238
|
zinc finger protein 238
|
|
chr3_-_98620452
|
0.473
|
NM_080927
|
DCBLD2
|
discoidin, CUB and LCCL domain containing 2
|
|
chr2_+_70142152
|
0.471
|
NM_001202513
NM_001202514
NM_002357
|
MXD1
|
MAX dimerization protein 1
|
|
chr3_+_143692166
|
0.467
|
NM_001134470
|
C3orf58
|
chromosome 3 open reading frame 58
|
|
chr9_-_98270830
|
0.465
|
NM_000264
|
PTCH1
|
patched 1
|
|
chr6_+_11538486
|
0.463
|
NM_001100829
|
TMEM170B
|
transmembrane protein 170B
|
|
chr12_+_111843743
|
0.456
|
NM_005475
|
SH2B3
|
SH2B adaptor protein 3
|
|
chr9_+_116638544
|
0.454
|
NM_133374
|
ZNF618
|
zinc finger protein 618
|
|
chr13_-_79233266
|
0.451
|
NM_024546
|
RNF219
|
ring finger protein 219
|
|
chr1_+_198125890
|
0.450
|
NM_133494
|
NEK7
|
NIMA (never in mitosis gene a)-related kinase 7
|
|
chr6_+_138483024
|
0.448
|
NM_020340
|
KIAA1244
|
KIAA1244
|
|
chr3_+_23987514
|
0.446
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr20_+_32150139
|
0.440
|
NM_001039709
NM_005093
|
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2
|
|
chr8_-_127570710
|
0.435
|
NM_174911
|
FAM84B
|
family with sequence similarity 84, member B
|
|
chr4_-_80994309
|
0.432
|
NM_001145794
NM_058172
|
ANTXR2
|
anthrax toxin receptor 2
|
|
chr7_+_8008416
|
0.423
|
NM_138426
|
GLCCI1
|
glucocorticoid induced transcript 1
|
|
chr11_-_74660165
|
0.422
|
NM_182969
|
XRRA1
|
X-ray radiation resistance associated 1
|
|
chr4_-_170192193
|
0.420
|
NM_020870
|
SH3RF1
|
SH3 domain containing ring finger 1
|
|
chr1_+_162466963
|
0.419
|
NM_001184763
|
UHMK1
|
U2AF homology motif (UHM) kinase 1
|
|
chr19_+_48111437
|
0.418
|
NM_015711
|
GLTSCR1
|
glioma tumor suppressor candidate region gene 1
|
|
chr3_+_170139027
|
0.417
|
NM_001185056
|
CLDN11
|
claudin 11
|
|
chr7_+_100026412
|
0.416
|
NM_001194990
NM_001194991
NM_001194992
|
MEPCE
|
methylphosphate capping enzyme
|
|
chr14_+_45431333
|
0.412
|
NM_015091
|
FAM179B
|
family with sequence similarity 179, member B
|
|
chr9_-_98269686
|
0.408
|
NM_001083606
|
PTCH1
|
patched 1
|
|
chr21_-_36421586
|
0.401
|
NM_001754
|
RUNX1
|
runt-related transcription factor 1
|
|
chr7_+_100027253
|
0.400
|
NM_019606
|
MEPCE
|
methylphosphate capping enzyme
|
|
chr9_-_98270321
|
0.398
|
NM_001083604
NM_001083605
|
PTCH1
|
patched 1
|
|
chr4_+_15004297
|
0.395
|
NM_001177381
NM_001177382
NM_001177383
NM_001177384
NM_182485
NM_182646
|
CPEB2
|
cytoplasmic polyadenylation element binding protein 2
|
|
chr3_+_107241782
|
0.395
|
NM_001142568
NM_020235
|
BBX
|
bobby sox homolog (Drosophila)
|
|
chr1_+_162467594
|
0.395
|
NM_144624
NM_175866
|
UHMK1
|
U2AF homology motif (UHM) kinase 1
|
|
chr18_+_8717368
|
0.395
|
NM_015210
|
CCDC165
|
coiled-coil domain containing 165
|
|
chr8_-_12990731
|
0.392
|
NM_006094
|
DLC1
|
deleted in liver cancer 1
|
|
chr10_+_18629613
|
0.391
|
NM_201590
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr10_-_105614952
|
0.385
|
NM_014631
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr1_-_156470506
|
0.384
|
NM_005920
|
MEF2D
|
myocyte enhancer factor 2D
|
|
chr12_-_76425375
|
0.382
|
NM_007350
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr10_-_124713693
|
0.378
|
NM_024942
|
C10orf88
|
chromosome 10 open reading frame 88
|
|
chr10_+_18429605
|
0.371
|
NM_201593
NM_201596
NM_201597
NM_001167945
NM_201571
NM_201572
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr3_+_143690639
|
0.371
|
NM_173552
|
C3orf58
|
chromosome 3 open reading frame 58
|
|
chr8_-_12973752
|
0.369
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr1_+_65730423
|
0.368
|
NM_014787
|
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6
|
|
chr17_-_65241305
|
0.368
|
NM_014877
|
HELZ
|
helicase with zinc finger
|
|
chr10_-_99393851
|
0.366
|
NM_001098831
|
MORN4
|
MORN repeat containing 4
|
|
chr1_+_116184573
|
0.365
|
NM_001172411
NM_138959
|
VANGL1
|
vang-like 1 (van gogh, Drosophila)
|
|
chr17_+_30469373
|
0.365
|
NM_001033566
NM_001033568
NM_018307
|
RHOT1
|
ras homolog gene family, member T1
|
|
chr7_-_107880492
|
0.364
|
NM_001037132
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr2_-_99279935
|
0.362
|
NM_001160154
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chr12_+_78225068
|
0.361
|
NM_014903
|
NAV3
|
neuron navigator 3
|
|
chr3_+_170136652
|
0.357
|
NM_005602
|
CLDN11
|
claudin 11
|
|
chr1_-_85725315
|
0.352
|
NM_198077
|
C1orf52
|
chromosome 1 open reading frame 52
|
|
chr5_+_137688270
|
0.352
|
NM_016604
|
KDM3B
|
lysine (K)-specific demethylase 3B
|
|
chr4_+_95679075
|
0.351
|
NM_001203
|
BMPR1B
|
bone morphogenetic protein receptor, type IB
|
|
chr16_+_58059281
|
0.351
|
NM_002428
|
MMP15
|
matrix metallopeptidase 15 (membrane-inserted)
|
|
chr5_+_139505519
|
0.350
|
NM_001007189
|
IGIP
|
IgA-inducing protein homolog (Bos taurus)
|
|
chr6_-_85473898
|
0.345
|
NM_001080508
|
TBX18
|
T-box 18
|
|
chr1_+_116185249
|
0.344
|
NM_001172412
|
VANGL1
|
vang-like 1 (van gogh, Drosophila)
|
|
chr11_-_95522945
|
0.342
|
NM_144664
|
FAM76B
|
family with sequence similarity 76, member B
|
|
chr10_+_60094712
|
0.341
|
NM_001204880
NM_003338
|
UBE2D1
|
ubiquitin-conjugating enzyme E2D 1
|
|
chr15_+_33603162
|
0.339
|
NM_001036
NM_001243996
|
RYR3
|
ryanodine receptor 3
|
|
chr10_+_74033676
|
0.338
|
NM_019058
|
DDIT4
|
DNA-damage-inducible transcript 4
|
|
chr5_+_153825497
|
0.333
|
NM_001131062
NM_001131063
NM_024632
|
SAP30L
|
SAP30-like
|
|
chr21_-_15755439
|
0.333
|
NM_006948
|
HSPA13
|
heat shock protein 70kDa family, member 13
|
|
chr20_+_32077904
|
0.329
|
NM_001032999
|
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2
|
|
chr9_-_98279246
|
0.327
|
NM_001083602
NM_001083603
|
PTCH1
|
patched 1
|
|
chr7_-_127670995
|
0.322
|
NM_022143
|
LRRC4
|
leucine rich repeat containing 4
|
|
chr6_+_163148163
|
0.321
|
NM_001080378
NM_152410
|
PACRG
|
PARK2 co-regulated
|
|
chr3_-_129612371
|
0.321
|
NM_001128224
|
TMCC1
|
transmembrane and coiled-coil domain family 1
|
|
chr2_-_99347588
|
0.318
|
NM_012214
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chr6_+_30524485
|
0.315
|
NM_001077497
NM_025263
|
PRR3
|
proline rich 3
|
|
chr12_+_4430346
|
0.312
|
NM_020375
|
C12orf5
|
chromosome 12 open reading frame 5
|
|
chr13_+_98605928
|
0.311
|
NM_002271
|
IPO5
|
importin 5
|
|
chr4_+_8271458
|
0.309
|
NM_053044
|
HTRA3
|
HtrA serine peptidase 3
|
|
chr16_-_89556886
|
0.305
|
NM_013275
|
ANKRD11
|
ankyrin repeat domain 11
|
|
chr22_+_46546498
|
0.302
|
NM_001001928
NM_005036
|
PPARA
|
peroxisome proliferator-activated receptor alpha
|
|
chr2_+_109065576
|
0.300
|
NM_181453
|
GCC2
|
GRIP and coiled-coil domain containing 2
|
|
chrX_+_144908927
|
0.296
|
NM_004709
|
CXorf1
|
chromosome X open reading frame 1
|
|
chr10_+_18689512
|
0.296
|
NM_201570
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr4_-_125633716
|
0.294
|
NM_001167882
NM_020337
|
ANKRD50
|
ankyrin repeat domain 50
|
|
chr16_+_88493878
|
0.293
|
NM_001127464
|
ZNF469
|
zinc finger protein 469
|
|
chr9_-_134151905
|
0.292
|
NM_033387
|
FAM78A
|
family with sequence similarity 78, member A
|
|
chr13_+_33160537
|
0.290
|
NM_015032
|
PDS5B
|
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae)
|
|
chr6_+_71377471
|
0.289
|
NM_001044305
NM_021940
|
SMAP1
|
small ArfGAP 1
|
|
chr22_+_47022657
|
0.288
|
NM_015124
|
GRAMD4
|
GRAM domain containing 4
|
|
chr12_+_121124922
|
0.287
|
NM_014730
|
MLEC
|
malectin
|
|
chr8_-_74884500
|
0.284
|
NM_001204857
NM_001204858
NM_001204859
NM_001204860
NM_001204863
NM_001204864
NM_005648
|
TCEB1
|
transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C)
|
|
chr17_+_36507708
|
0.282
|
NM_014598
|
SOCS7
|
suppressor of cytokine signaling 7
|
|
chr3_-_69101236
|
0.282
|
NM_007114
|
TMF1
|
TATA element modulatory factor 1
|
|
chr2_+_60983354
|
0.280
|
NM_022894
|
PAPOLG
|
poly(A) polymerase gamma
|
|
chr5_-_40798263
|
0.276
|
NM_006251
NM_206907
|
PRKAA1
|
protein kinase, AMP-activated, alpha 1 catalytic subunit
|
|
chr15_+_84322809
|
0.275
|
NM_207517
|
ADAMTSL3
|
ADAMTS-like 3
|
|
chr14_-_52535945
|
0.275
|
NM_007361
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr5_+_115177616
|
0.274
|
NM_001284
|
AP3S1
|
adaptor-related protein complex 3, sigma 1 subunit
|
|
chr8_-_74884142
|
0.274
|
NM_001204861
NM_001204862
|
TCEB1
|
transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C)
|
|
chr2_+_198381021
|
0.272
|
NM_001204094
|
MOB4
|
MOB family member 4, phocein
|
|
chr12_-_131002409
|
0.271
|
NM_015347
|
RIMBP2
|
RIMS binding protein 2
|
|
chr3_+_23986735
|
0.271
|
NM_005126
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr4_+_26862312
|
0.271
|
NM_001169117
NM_001169118
NM_020860
|
STIM2
|
stromal interaction molecule 2
|
|
chr5_-_107006585
|
0.270
|
NM_001962
|
EFNA5
|
ephrin-A5
|
|
chr10_+_98592015
|
0.269
|
NM_001170765
NM_001170766
|
LCOR
|
ligand dependent nuclear receptor corepressor
|
|
chr5_-_126366439
|
0.268
|
NM_178450
|
MARCH3
|
membrane-associated ring finger (C3HC4) 3
|
|
chrX_-_109683457
|
0.267
|
NM_001171689
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chrX_-_33229428
|
0.266
|
NM_004006
|
DMD
|
dystrophin
|
|
chr3_-_129599220
|
0.266
|
NM_001017395
|
TMCC1
|
transmembrane and coiled-coil domain family 1
|
|
chr1_+_169075869
|
0.266
|
NM_001677
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr2_+_74056095
|
0.266
|
NM_006463
NM_213622
|
STAMBP
|
STAM binding protein
|
|
chr20_-_52199635
|
0.264
|
NM_006526
|
ZNF217
|
zinc finger protein 217
|
|
chr22_+_40573879
|
0.264
|
NM_001162501
NM_015088
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr1_-_72748147
|
0.261
|
NM_173808
|
NEGR1
|
neuronal growth regulator 1
|
|
chr3_+_67048726
|
0.261
|
NM_032505
|
KBTBD8
|
kelch repeat and BTB (POZ) domain containing 8
|
|
chr11_-_34379526
|
0.257
|
NM_145804
|
ABTB2
|
ankyrin repeat and BTB (POZ) domain containing 2
|
|
chr6_+_163149001
|
0.255
|
NM_001080379
|
PACRG
|
PARK2 co-regulated
|
|
chr6_-_111804413
|
0.252
|
NM_002912
|
REV3L
|
REV3-like, catalytic subunit of DNA polymerase zeta (yeast)
|
|
chr19_+_10654591
|
0.252
|
NM_032885
|
ATG4D
|
ATG4 autophagy related 4 homolog D (S. cerevisiae)
|
|
chr7_-_105029321
|
0.251
|
NM_182692
|
SRPK2
|
SRSF protein kinase 2
|
|
chr7_-_149194896
|
0.250
|
NM_001163474
NM_152557
|
ZNF746
|
zinc finger protein 746
|
|
chrX_-_109561314
|
0.250
|
NM_001025580
NM_015365
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr17_-_27169740
|
0.247
|
NM_001077498
NM_018182
|
C17orf63
|
chromosome 17 open reading frame 63
|
|
chrX_-_108976509
|
0.247
|
NM_004458
NM_022977
|
ACSL4
|
acyl-CoA synthetase long-chain family member 4
|
|
chr12_+_69004614
|
0.247
|
NM_001010942
NM_001251917
NM_001251918
NM_001251921
NM_001251922
NM_015646
|
RAP1B
|
RAP1B, member of RAS oncogene family
|
|
chr14_+_93673400
|
0.244
|
NM_175748
|
UBR7
|
ubiquitin protein ligase E3 component n-recognin 7 (putative)
|
|
chr8_-_95961543
|
0.242
|
NM_001135733
NM_033285
|
TP53INP1
|
tumor protein p53 inducible nuclear protein 1
|
|
chrX_-_31526414
|
0.242
|
NM_004014
|
DMD
|
dystrophin
|
|
chr2_+_198380294
|
0.242
|
NM_199482
|
MOB4
|
MOB family member 4, phocein
|
|
chr9_-_6015606
|
0.241
|
NM_001243202
NM_001243203
NM_012416
|
RANBP6
|
RAN binding protein 6
|
|
chr12_-_76478737
|
0.241
|
NM_004537
NM_139207
|
NAP1L1
|
nucleosome assembly protein 1-like 1
|
|
chr5_+_148520985
|
0.238
|
NM_014945
|
ABLIM3
|
actin binding LIM protein family, member 3
|
|
chr7_-_104909442
|
0.237
|
NM_182691
|
SRPK2
|
SRSF protein kinase 2
|
|
chr14_-_27066959
|
0.235
|
NM_002515
NM_006489
NM_006491
|
NOVA1
|
neuro-oncological ventral antigen 1
|