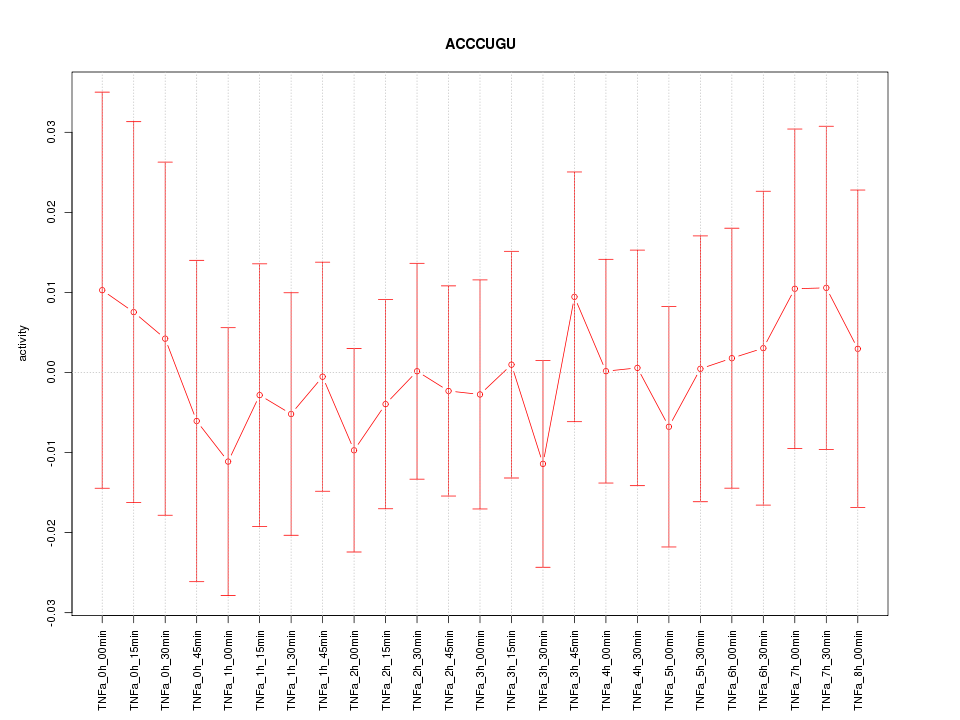
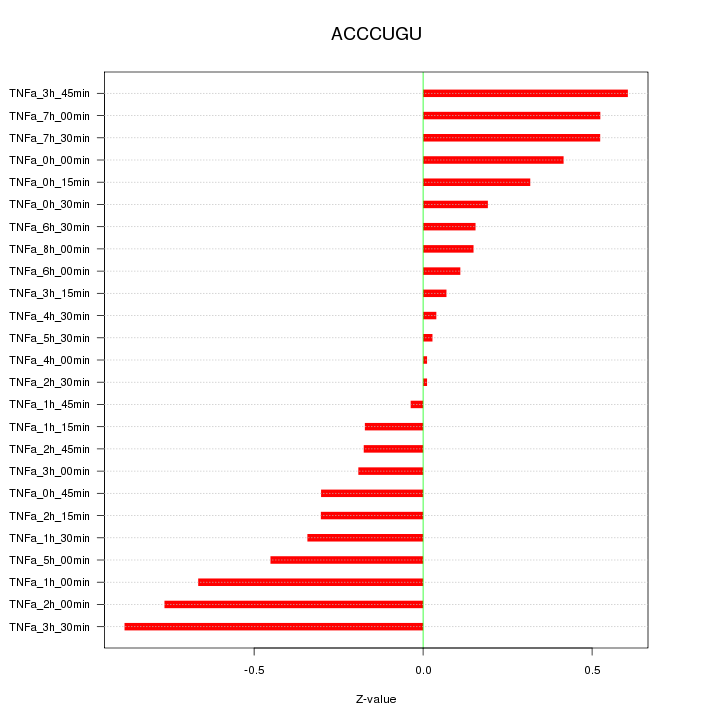
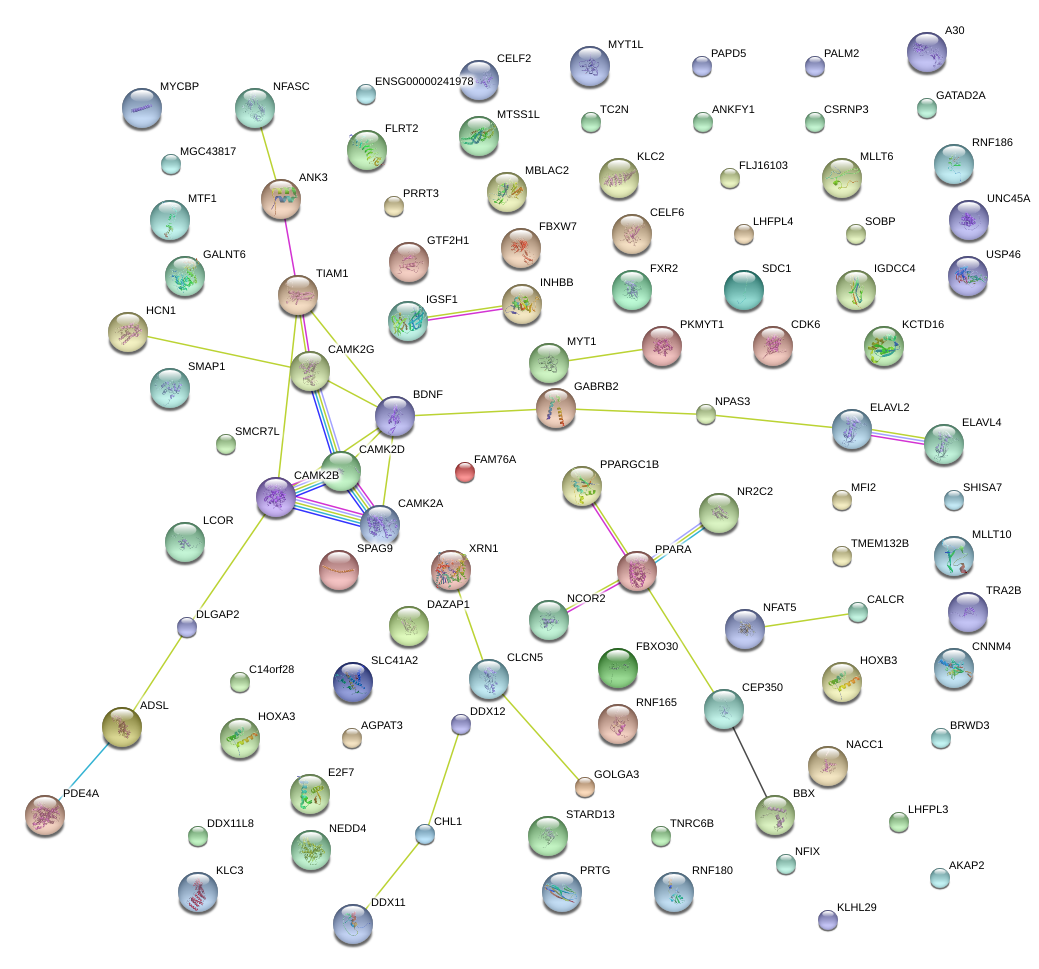
|
chr13_-_33760215
|
0.464
|
NM_001243466
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr13_-_33780142
|
0.454
|
NM_001243474
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr13_-_33859835
|
0.453
|
NM_178006
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr5_+_143550395
|
0.423
|
NM_020768
|
KCTD16
|
potassium channel tetramerisation domain containing 16
|
|
chr16_+_50186809
|
0.403
|
NM_001040284
NM_001040285
|
PAPD5
|
PAP associated domain containing 5
|
|
chr3_-_9994057
|
0.366
|
NM_207351
|
PRRT3
|
proline-rich transmembrane protein 3
|
|
chr7_-_27159213
|
0.358
|
NM_030661
|
HOXA3
|
homeobox A3
|
|
chr12_-_105322056
|
0.350
|
NM_032148
|
SLC41A2
|
solute carrier family 41, member 2
|
|
chr11_-_27721975
|
0.349
|
NM_001143813
NM_001143814
NM_001709
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_-_27722599
|
0.345
|
NM_001143808
NM_001143809
NM_001143810
NM_001143811
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_-_27722446
|
0.345
|
NM_001143812
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr2_+_23608297
|
0.343
|
NM_052920
|
KLHL29
|
kelch-like 29 (Drosophila)
|
|
chr11_-_27743604
|
0.341
|
NM_170731
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr7_-_27166638
|
0.338
|
NM_153631
|
HOXA3
|
homeobox A3
|
|
chr11_-_27723061
|
0.337
|
NM_170733
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_-_27741192
|
0.321
|
NM_001143807
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_-_27742224
|
0.312
|
NM_001143805
NM_001143806
NM_170732
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_-_27721179
|
0.311
|
NM_170734
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_-_27681195
|
0.303
|
NM_001143816
NM_170735
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr10_-_62149487
|
0.248
|
NM_020987
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr5_+_149109814
|
0.226
|
NM_001172698
NM_133263
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr1_-_39338976
|
0.210
|
NM_012333
|
MYCBP
|
c-myc binding protein
|
|
chr5_+_149151502
|
0.203
|
NM_001172699
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr10_-_62493282
|
0.203
|
NM_001204403
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr10_-_61900659
|
0.200
|
NM_001149
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr9_-_23821477
|
0.176
|
NM_001171197
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr19_+_10543110
|
0.168
|
NM_001111309
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr19_+_10531128
|
0.167
|
NM_001111307
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr2_+_166428845
|
0.167
|
NM_024969
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr10_-_62332666
|
0.166
|
NM_001204404
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr18_+_43914186
|
0.164
|
NM_152470
|
RNF165-IT1
RNF165
|
RNF165 intronic transcript 1 (non-protein coding)
ring finger protein 165
|
|
chr9_-_23821842
|
0.159
|
NM_001171195
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr19_+_10541334
|
0.158
|
NM_001111308
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr9_-_23826062
|
0.154
|
NM_004432
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr2_+_166326156
|
0.148
|
NM_001172173
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chrX_+_49832214
|
0.141
|
NM_000084
|
CLCN5
|
chloride channel 5
|
|
chr2_-_2334887
|
0.136
|
NM_015025
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr22_+_40440664
|
0.133
|
NM_001024843
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr15_-_56035176
|
0.130
|
NM_173814
|
PRTG
|
protogenin
|
|
chr12_-_125052009
|
0.129
|
NM_001077261
NM_001206654
NM_006312
|
NCOR2
|
nuclear receptor corepressor 2
|
|
chr19_+_13105970
|
0.126
|
NM_002501
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr17_-_46651809
|
0.124
|
NM_002146
|
HOXB3
|
homeobox B3
|
|
chr19_+_10563581
|
0.116
|
NM_006202
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr8_+_1449531
|
0.115
|
NM_004745
|
DLGAP2
|
discs, large (Drosophila) homolog-associated protein 2
|
|
chr15_-_65715385
|
0.111
|
NM_020962
|
IGDCC4
|
immunoglobulin superfamily, DCC subclass, member 4
|
|
chr1_+_204913353
|
0.107
|
NM_001160331
|
NFASC
|
neurofascin
|
|
chr5_-_160975124
|
0.106
|
NM_000813
NM_021911
|
GABRB2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2
|
|
chr17_-_49198084
|
0.102
|
NM_001130527
NM_001130528
NM_003971
|
SPAG9
|
sperm associated antigen 9
|
|
chr19_+_19496629
|
0.100
|
NM_017660
|
GATAD2A
|
GATA zinc finger domain containing 2A
|
|
chr3_-_142166782
|
0.097
|
NM_001042604
NM_019001
|
XRN1
|
5'-3' exoribonuclease 1
|
|
chr22_+_46546498
|
0.095
|
NM_001001928
NM_005036
|
PPARA
|
peroxisome proliferator-activated receptor alpha
|
|
chr3_-_9595412
|
0.092
|
NM_198560
|
LHFPL4
|
lipoma HMGIC fusion partner-like 4
|
|
chr12_-_77459198
|
0.087
|
NM_203394
|
E2F7
|
E2F transcription factor 7
|
|
chr14_+_45366426
|
0.085
|
NM_001017923
|
C14orf28
|
chromosome 14 open reading frame 28
|
|
chr17_-_4167028
|
0.082
|
NM_016376
NM_020740
|
ANKFY1
|
ankyrin repeat and FYVE domain containing 1
|
|
chr14_+_85996454
|
0.080
|
NM_013231
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr19_-_55954229
|
0.080
|
NM_001145176
|
SHISA7
|
shisa homolog 7 (Xenopus laevis)
|
|
chr10_+_11047253
|
0.079
|
NM_001025077
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr19_+_13229101
|
0.077
|
NM_052876
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing
|
|
chr1_+_179923870
|
0.077
|
NM_014810
|
CEP350
|
centrosomal protein 350kDa
|
|
chr1_+_28052467
|
0.077
|
NM_001143912
NM_001143913
NM_001143914
NM_001143915
NM_152660
|
FAM76A
|
family with sequence similarity 76, member A
|
|
chr7_-_44365019
|
0.076
|
NM_001220
NM_172078
NM_172079
NM_172080
NM_172081
NM_172082
NM_172083
NM_172084
|
CAMK2B
|
calcium/calmodulin-dependent protein kinase II beta
|
|
chr19_+_1407531
|
0.073
|
NM_018959
NM_170711
|
DAZAP1
|
DAZ associated protein 1
|
|
chr3_-_185655833
|
0.070
|
NM_001243879
NM_004593
|
TRA2B
|
transformer 2 beta homolog (Drosophila)
|
|
chrX_-_130423034
|
0.070
|
NM_001170963
|
IGSF1
|
immunoglobulin superfamily, member 1
|
|
chr9_+_112403067
|
0.070
|
NM_001037293
|
PALM2
|
paralemmin 2
|
|
chr11_+_66025070
|
0.069
|
NM_001134776
NM_001134774
NM_022822
|
KLC2
|
kinesin light chain 2
|
|
chr7_-_93204035
|
0.067
|
NM_001164737
NM_001742
|
CALCR
|
calcitonin receptor
|
|
chr21_+_45285101
|
0.067
|
NM_020132
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3
|
|
chr15_-_56285732
|
0.066
|
NM_006154
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chrX_+_49687212
|
0.065
|
NM_001127898
NM_001127899
|
CLCN5
|
chloride channel 5
|
|
chr5_-_45696219
|
0.064
|
NM_021072
|
HCN1
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 1
|
|
chr16_-_70719854
|
0.064
|
NM_138383
|
MTSS1L
|
metastasis suppressor 1-like
|
|
chr22_+_40573879
|
0.064
|
NM_001162501
NM_015088
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr2_+_97426571
|
0.063
|
NM_020184
|
CNNM4
|
cyclin M4
|
|
chr3_+_238274
|
0.063
|
NM_001253387
NM_006614
|
CHL1
|
cell adhesion molecule with homology to L1CAM (close homolog of L1)
|
|
chr5_-_89770432
|
0.059
|
NM_203406
|
MBLAC2
|
metallo-beta-lactamase domain containing 2
|
|
chr11_+_66024746
|
0.056
|
NM_001134775
|
KLC2
|
kinesin light chain 2
|
|
chr7_-_93189121
|
0.055
|
NM_001164738
|
CALCR
|
calcitonin receptor
|
|
chr1_+_204797774
|
0.055
|
NM_001005388
NM_001005389
NM_001160332
NM_001160333
NM_015090
|
NFASC
|
neurofascin
|
|
chr17_-_7518124
|
0.054
|
NM_004860
|
FXR2
|
fragile X mental retardation, autosomal homolog 2
|
|
chr2_-_20424627
|
0.053
|
NM_002997
|
SDC1
|
syndecan 1
|
|
chr7_-_92463211
|
0.052
|
NM_001259
|
CDK6
|
cyclin-dependent kinase 6
|
|
chr6_-_146135918
|
0.052
|
NM_032145
|
FBXO30
|
F-box protein 30
|
|
chr3_+_107241782
|
0.050
|
NM_001142568
NM_020235
|
BBX
|
bobby sox homolog (Drosophila)
|
|
chrX_-_80065182
|
0.050
|
NM_153252
|
BRWD3
|
bromodomain and WD repeat domain containing 3
|
|
chr12_+_125811161
|
0.048
|
NM_052907
|
TMEM132B
|
transmembrane protein 132B
|
|
chr21_+_45345278
|
0.048
|
NM_001037553
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3
|
|
chr4_-_53522756
|
0.045
|
NM_001134223
|
USP46
|
ubiquitin specific peptidase 46
|
|
chr12_-_133405197
|
0.044
|
NM_005895
|
GOLGA3
|
golgin A3
|
|
chr14_+_33408458
|
0.044
|
NM_001164749
NM_001165893
NM_022123
NM_173159
|
NPAS3
|
neuronal PAS domain protein 3
|
|
chr7_-_92465929
|
0.043
|
NM_001145306
|
CDK6
|
cyclin-dependent kinase 6
|
|
chr6_+_107811272
|
0.043
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr22_+_39898248
|
0.041
|
NM_019008
|
SMCR7L
|
Smith-Magenis syndrome chromosome region, candidate 7-like
|
|
chr15_-_56209305
|
0.041
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr21_-_32931289
|
0.041
|
NM_003253
|
TIAM1
|
T-cell lymphoma invasion and metastasis 1
|
|
chr10_+_98592658
|
0.041
|
NM_032440
|
LCOR
|
ligand dependent nuclear receptor corepressor
|
|
chr4_-_53525316
|
0.040
|
NM_022832
|
USP46
|
ubiquitin specific peptidase 46
|
|
chr1_-_38325224
|
0.040
|
NM_005955
|
MTF1
|
metal-regulatory transcription factor 1
|
|
chr1_-_20141593
|
0.037
|
NM_019062
|
RNF186
|
ring finger protein 186
|
|
chr2_-_20425166
|
0.037
|
NM_001006946
|
SDC1
|
syndecan 1
|
|
chr11_+_18343794
|
0.034
|
NM_001142307
NM_005316
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa
|
|
chr16_+_69599868
|
0.033
|
NM_001113178
NM_006599
NM_138713
NM_138714
NM_173214
NM_173215
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr3_+_14989076
|
0.033
|
NM_003298
|
NR2C2
|
nuclear receptor subfamily 2, group C, member 2
|
|
chr6_+_71377471
|
0.033
|
NM_001044305
NM_021940
|
SMAP1
|
small ArfGAP 1
|
|
chr15_-_72612497
|
0.032
|
NM_052840
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr15_-_72612247
|
0.031
|
NM_001172684
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr17_+_36861857
|
0.031
|
NM_005937
|
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6
|
|
chr1_-_38230804
|
0.031
|
NM_001099439
NM_173641
|
EPHA10
|
EPH receptor A10
|
|
chr5_+_63461652
|
0.029
|
NM_001113561
NM_178532
|
RNF180
|
ring finger protein 180
|
|
chr2_+_121103670
|
0.028
|
NM_002193
|
INHBB
|
inhibin, beta B
|
|
chr9_+_129567260
|
0.028
|
NM_001135776
NM_014007
|
ZBTB43
|
zinc finger and BTB domain containing 43
|
|
chr18_+_9136742
|
0.027
|
NM_001083625
NM_015208
|
ANKRD12
|
ankyrin repeat domain 12
|
|
chr1_+_57110745
|
0.027
|
NM_006252
|
PRKAA2
|
protein kinase, AMP-activated, alpha 2 catalytic subunit
|
|
chr11_-_8680382
|
0.026
|
NM_014818
|
TRIM66
|
tripartite motif containing 66
|
|
chr10_-_75173807
|
0.026
|
NM_001156
NM_004034
|
ANXA7
|
annexin A7
|
|
chr12_-_114846191
|
0.025
|
NM_000192
NM_080717
|
TBX5
|
T-box 5
|
|
chr7_-_72936592
|
0.025
|
NM_032408
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B
|
|
chr2_-_37193609
|
0.024
|
NM_003162
|
STRN
|
striatin, calmodulin binding protein
|
|
chr17_-_1359410
|
0.023
|
NM_005206
NM_016823
|
CRK
|
v-crk sarcoma virus CT10 oncogene homolog (avian)
|
|
chr4_+_184020343
|
0.023
|
NM_024949
|
WWC2
|
WW and C2 domain containing 2
|
|
chrX_-_73834460
|
0.020
|
NM_016120
NM_183353
|
RLIM
|
ring finger protein, LIM domain interacting
|
|
chr7_-_5463176
|
0.019
|
NM_001080495
|
TNRC18
|
trinucleotide repeat containing 18
|
|
chr18_+_9137560
|
0.019
|
NM_001204056
|
ANKRD12
|
ankyrin repeat domain 12
|
|
chrX_-_129402753
|
0.019
|
NM_017666
|
ZNF280C
|
zinc finger protein 280C
|
|
chr2_-_128100673
|
0.019
|
NM_006609
|
MAP3K2
|
mitogen-activated protein kinase kinase kinase 2
|
|
chr6_+_116421970
|
0.017
|
NM_152729
|
NT5DC1
|
5'-nucleotidase domain containing 1
|
|
chr20_+_55204307
|
0.017
|
NM_003222
|
TFAP2C
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma)
|
|
chr22_+_45560529
|
0.016
|
NM_153645
|
NUP50
|
nucleoporin 50kDa
|
|
chr8_-_25902639
|
0.015
|
NM_022659
|
EBF2
|
early B-cell factor 2
|
|
chr9_+_100263461
|
0.014
|
NM_001166116
|
TMOD1
|
tropomodulin 1
|
|
chr22_+_45559725
|
0.014
|
NM_007172
|
NUP50
|
nucleoporin 50kDa
|
|
chr12_-_85306579
|
0.013
|
NM_001146335
NM_018057
NM_182767
|
SLC6A15
|
solute carrier family 6 (neutral amino acid transporter), member 15
|
|
chr17_-_56084656
|
0.013
|
NM_001078166
NM_006924
|
SRSF1
|
serine/arginine-rich splicing factor 1
|
|
chr15_-_72598655
|
0.013
|
NM_001172685
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr14_-_47812394
|
0.012
|
NM_182830
|
MDGA2
|
MAM domain containing glycosylphosphatidylinositol anchor 2
|
|
chr3_-_196159304
|
0.012
|
NM_015562
|
UBXN7
|
UBX domain protein 7
|
|
chr12_-_22487566
|
0.011
|
NM_003034
|
ST8SIA1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1
|
|
chr10_-_128994421
|
0.011
|
NM_001039762
|
FAM196A
|
family with sequence similarity 196, member A
|
|
chr17_+_18218569
|
0.010
|
NM_144775
|
SMCR8
|
Smith-Magenis syndrome chromosome region, candidate 8
|
|
chr14_+_99947732
|
0.010
|
NM_001099402
|
CCNK
|
cyclin K
|
|
chr8_-_16050220
|
0.009
|
NM_002445
NM_138715
NM_138716
|
MSR1
|
macrophage scavenger receptor 1
|
|
chr10_-_88281515
|
0.009
|
NM_015045
|
WAPAL
|
wings apart-like homolog (Drosophila)
|
|
chr15_-_60884615
|
0.007
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr12_-_54673901
|
0.006
|
NM_012117
|
CBX5
|
chromobox homolog 5
|
|
chr1_+_26758772
|
0.005
|
NM_001243564
NM_001243565
NM_024887
NM_205861
|
DHDDS
|
dehydrodolichyl diphosphate synthase
|
|
chr17_-_79479831
|
0.004
|
NM_001199954
NM_001614
|
ACTG1
|
actin, gamma 1
|
|
chr1_+_199996729
|
0.004
|
NM_003822
NM_205860
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2
|
|
chr12_+_4918341
|
0.003
|
NM_002235
|
KCNA6
|
potassium voltage-gated channel, shaker-related subfamily, member 6
|
|
chr3_+_48956272
|
0.002
|
NM_006321
|
ARIH2
|
ariadne homolog 2 (Drosophila)
|
|
chr12_-_54653317
|
0.002
|
NM_001127322
|
CBX5
|
chromobox homolog 5
|
|
chr19_-_56110851
|
0.001
|
NM_032836
|
FIZ1
|
FLT3-interacting zinc finger 1
|
|
chr3_+_183873253
|
0.001
|
NM_004423
|
DVL3
|
dishevelled, dsh homolog 3 (Drosophila)
|