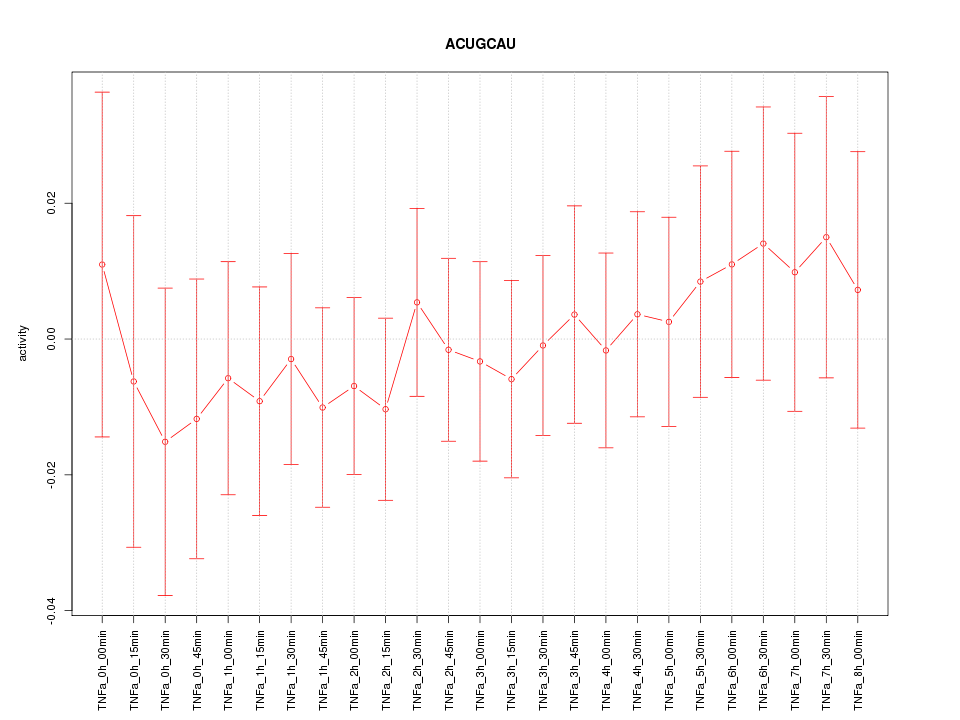
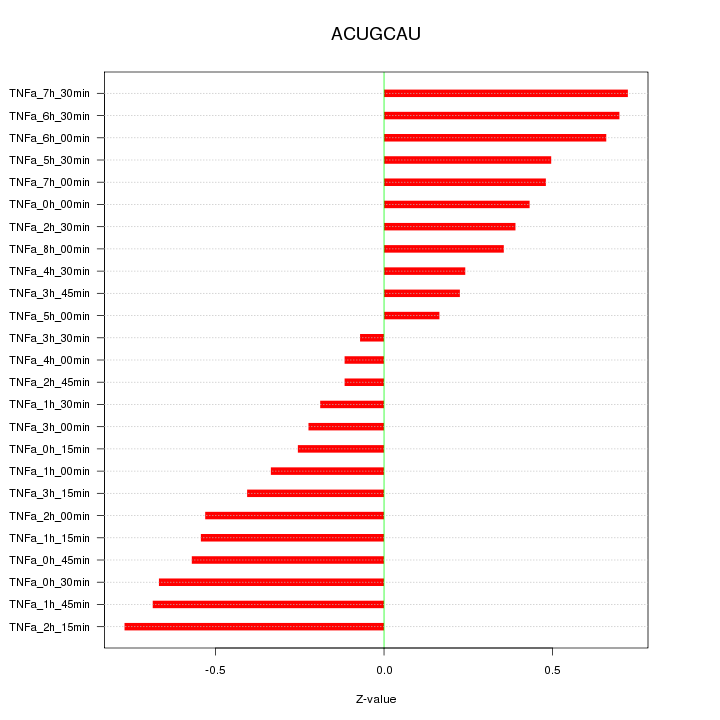
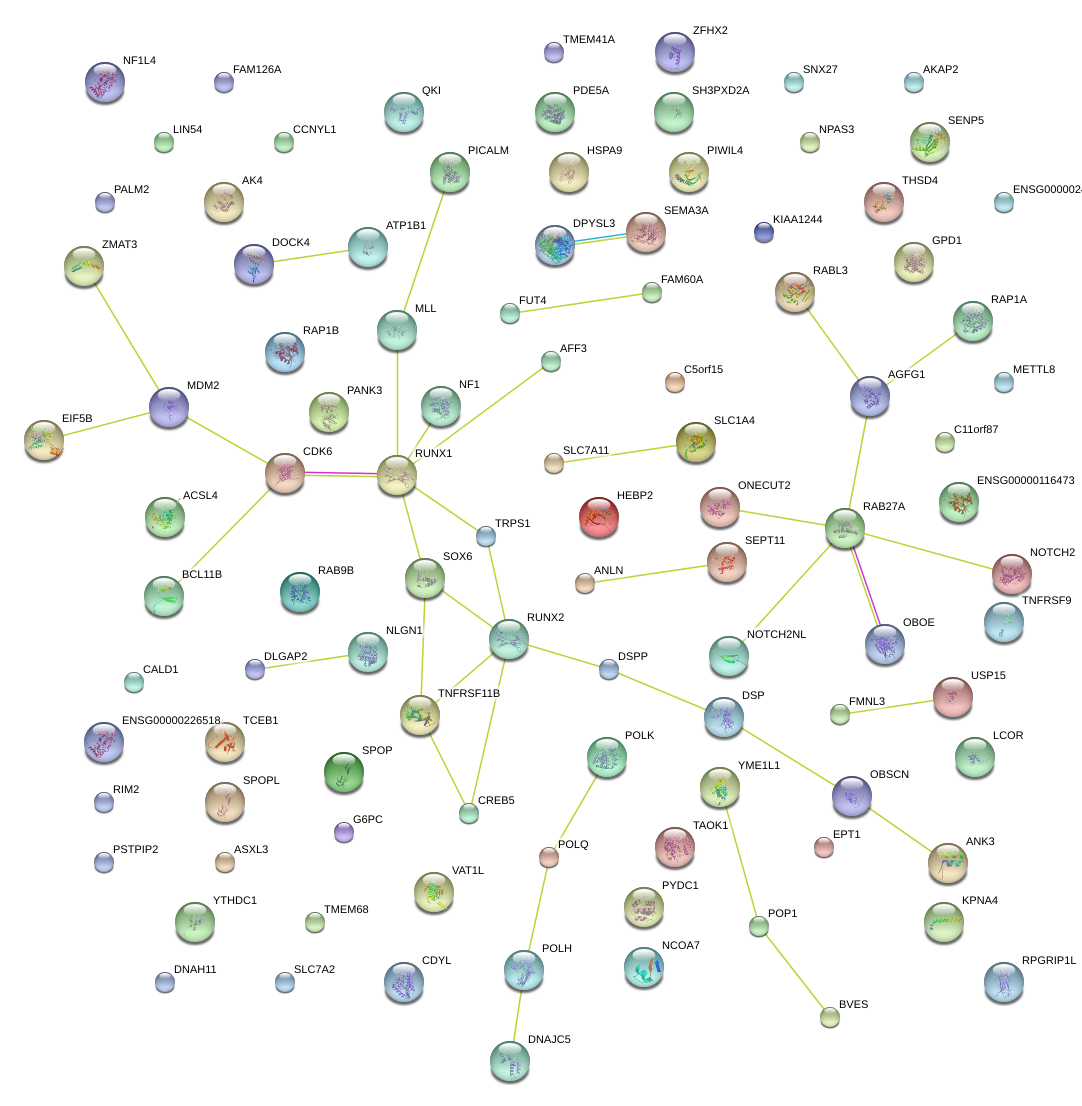
|
chr2_-_100721792
|
1.148
|
NM_001025108
|
AFF3
|
AF4/FMR2 family, member 3
|
|
chr10_-_105614952
|
1.142
|
NM_014631
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr2_-_100758970
|
0.981
|
NM_002285
|
AFF3
|
AF4/FMR2 family, member 3
|
|
chr7_-_83824216
|
0.518
|
NM_006080
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A
|
|
chr21_-_36421586
|
0.451
|
NM_001754
|
RUNX1
|
runt-related transcription factor 1
|
|
chr3_-_160283361
|
0.390
|
NM_002268
|
KPNA4
|
karyopherin alpha 4 (importin alpha 3)
|
|
chr3_+_173116243
|
0.382
|
NM_014932
|
NLGN1
|
neuroligin 1
|
|
chr18_-_43652153
|
0.376
|
NM_024430
|
PSTPIP2
|
proline-serine-threonine phosphatase interacting protein 2
|
|
chr8_+_17354562
|
0.367
|
NM_001008539
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr10_-_62149487
|
0.360
|
NM_020987
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr8_+_17396285
|
0.352
|
NM_001164771
NM_003046
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr1_-_8003224
|
0.348
|
NM_001561
|
TNFRSF9
|
tumor necrosis factor receptor superfamily, member 9
|
|
chr4_+_77870864
|
0.336
|
NM_018243
|
SEPT11
|
septin 11
|
|
chr4_-_120549115
|
0.320
|
NM_033437
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific
|
|
chr5_-_137911317
|
0.282
|
NM_004134
|
HSPA9
|
heat shock 70kDa protein 9 (mortalin)
|
|
chr10_-_61900659
|
0.279
|
NM_001149
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr10_-_62493282
|
0.279
|
NM_001204403
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr20_+_62526453
|
0.265
|
NM_025219
|
DNAJC5
|
DnaJ (Hsp40) homolog, subfamily C, member 5
|
|
chr1_+_65613771
|
0.260
|
NM_013410
|
AK4
|
adenylate kinase 4
|
|
chr1_+_169075869
|
0.259
|
NM_001677
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr1_+_65613211
|
0.255
|
NM_203464
NM_001005353
|
AK4
|
adenylate kinase 4
|
|
chr10_-_62332666
|
0.245
|
NM_001204404
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr10_+_98592658
|
0.240
|
NM_032440
|
LCOR
|
ligand dependent nuclear receptor corepressor
|
|
chr4_-_120549827
|
0.229
|
NM_001083
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific
|
|
chr9_+_112810877
|
0.202
|
NM_001004065
NM_001198656
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chr16_+_77822400
|
0.198
|
NM_020927
|
VAT1L
|
vesicle amine transport protein 1 homolog (T. californica)-like
|
|
chr5_-_146833150
|
0.196
|
NM_001387
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr10_+_98592015
|
0.194
|
NM_001170765
NM_001170766
|
LCOR
|
ligand dependent nuclear receptor corepressor
|
|
chr5_-_133304274
|
0.191
|
NM_020199
|
C5orf15
|
chromosome 5 open reading frame 15
|
|
chr1_+_151584514
|
0.189
|
NM_030918
|
SNX27
|
sorting nexin family member 27
|
|
chr6_+_126111782
|
0.186
|
NM_001122842
NM_001199621
NM_181782
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr4_-_120548329
|
0.186
|
NM_033430
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific
|
|
chr8_-_119964059
|
0.181
|
NM_002546
|
TNFRSF11B
|
tumor necrosis factor receptor superfamily, member 11b
|
|
chr14_+_33408458
|
0.180
|
NM_001164749
NM_001165893
NM_022123
NM_173159
|
NPAS3
|
neuronal PAS domain protein 3
|
|
chr1_+_145209067
|
0.177
|
NM_203458
|
NOTCH2NL
|
notch 2 N-terminal like
|
|
chr9_+_112887780
|
0.172
|
NM_001136562
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chr2_+_208576239
|
0.171
|
NM_001142300
NM_152523
|
CCNYL1
|
cyclin Y-like 1
|
|
chr8_+_104512975
|
0.166
|
NM_001100117
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr4_-_139163222
|
0.166
|
NM_014331
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11
|
|
chr8_-_56685869
|
0.165
|
NM_152417
|
TMEM68
|
transmembrane protein 68
|
|
chr18_+_31158540
|
0.165
|
NM_030632
|
ASXL3
|
additional sex combs like 3 (Drosophila)
|
|
chr21_-_36260972
|
0.165
|
NM_001001890
NM_001122607
|
RUNX1
|
runt-related transcription factor 1
|
|
chr3_-_120461362
|
0.165
|
NM_173825
|
RABL3
|
RAB, member of RAS oncogene family-like 3
|
|
chr16_-_53737713
|
0.164
|
NM_001127897
NM_015272
|
RPGRIP1L
|
RPGRIP1-like
|
|
chr12_+_62654120
|
0.163
|
NM_001252078
NM_001252079
NM_006313
|
USP15
|
ubiquitin specific peptidase 15
|
|
chr15_+_71433787
|
0.161
|
NM_024817
|
THSD4
|
thrombospondin, type I, domain containing 4
|
|
chr7_+_36429388
|
0.156
|
NM_018685
|
ANLN
|
anillin, actin binding protein
|
|
chr10_-_27443309
|
0.155
|
NM_001253866
NM_014263
NM_139312
|
YME1L1
|
YME1-like 1 (S. cerevisiae)
|
|
chr6_+_4776641
|
0.153
|
NM_004824
|
CDYL
|
chromodomain protein, Y-like
|
|
chr6_+_4890225
|
0.151
|
NM_001143971
|
CDYL
|
chromodomain protein, Y-like
|
|
chrX_-_108976509
|
0.148
|
NM_004458
NM_022977
|
ACSL4
|
acyl-CoA synthetase long-chain family member 4
|
|
chr5_-_146889618
|
0.147
|
NM_001197294
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr2_+_99953749
|
0.145
|
NM_015904
|
EIF5B
|
eukaryotic translation initiation factor 5B
|
|
chr5_-_168006413
|
0.144
|
NM_024594
|
PANK3
|
pantothenate kinase 3
|
|
chr6_+_4836331
|
0.144
|
NM_001143970
|
CDYL
|
chromodomain protein, Y-like
|
|
chr8_+_104831415
|
0.135
|
NM_014677
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr11_+_109292837
|
0.134
|
NM_207645
|
C11orf87
|
chromosome 11 open reading frame 87
|
|
chr17_+_41052813
|
0.133
|
NM_000151
|
G6PC
|
glucose-6-phosphatase, catalytic subunit
|
|
chr11_+_94277005
|
0.133
|
NM_002033
|
PIWIL4
FUT4
|
piwi-like 4 (Drosophila)
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific)
|
|
chr7_+_28725597
|
0.133
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr8_+_1449531
|
0.132
|
NM_004745
|
DLGAP2
|
discs, large (Drosophila) homolog-associated protein 2
|
|
chr11_-_16424391
|
0.132
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr7_+_28452143
|
0.132
|
NM_182898
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr6_+_7541863
|
0.130
|
NM_001008844
NM_004415
|
DSP
|
desmoplakin
|
|
chr2_+_65216482
|
0.129
|
NM_003038
|
SLC1A4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4
|
|
chr12_-_50101192
|
0.129
|
NM_175736
NM_198900
|
FMNL3
|
formin-like 3
|
|
chr2_+_65215577
|
0.127
|
NM_001193493
|
SLC1A4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4
|
|
chr2_+_228336877
|
0.126
|
NM_001135187
NM_001135188
NM_001135189
NM_004504
|
AGFG1
|
ArfGAP with FG repeats 1
|
|
chr2_+_26568900
|
0.122
|
NM_033505
|
EPT1
|
ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific)
|
|
chr15_-_55562483
|
0.121
|
NM_183234
NM_004580
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr7_-_92465929
|
0.121
|
NM_001145306
|
CDK6
|
cyclin-dependent kinase 6
|
|
chr7_+_134464161
|
0.119
|
NM_004342
NM_033138
NM_033157
|
CALD1
|
caldesmon 1
|
|
chr6_+_126240369
|
0.118
|
NM_001199622
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr11_+_118307072
|
0.118
|
NM_001197104
NM_005933
|
MLL
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila)
|
|
chr3_-_178789597
|
0.117
|
NM_022470
NM_152240
|
ZMAT3
|
zinc finger, matrin-type 3
|
|
chr7_-_92463211
|
0.116
|
NM_001259
|
CDK6
|
cyclin-dependent kinase 6
|
|
chr3_-_185216545
|
0.116
|
NM_080652
|
TMEM41A
|
transmembrane protein 41A
|
|
chr7_-_23053732
|
0.115
|
NM_032581
|
FAM126A
|
family with sequence similarity 126, member A
|
|
chr11_-_85780105
|
0.114
|
NM_001008660
NM_001206946
NM_007166
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr15_-_55563091
|
0.114
|
NM_183236
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr8_-_74884142
|
0.113
|
NM_001204861
NM_001204862
|
TCEB1
|
transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C)
|
|
chr17_+_27717942
|
0.112
|
NM_020791
NM_025142
|
TAOK1
|
TAO kinase 1
|
|
chr8_-_74884500
|
0.109
|
NM_001204857
NM_001204858
NM_001204859
NM_001204860
NM_001204863
NM_001204864
NM_005648
|
TCEB1
|
transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C)
|
|
chr12_+_69201845
|
0.108
|
NM_002392
|
MDM2
|
Mdm2 p53 binding protein homolog (mouse)
|
|
chr12_+_69004614
|
0.108
|
NM_001010942
NM_001251917
NM_001251918
NM_001251921
NM_001251922
NM_015646
|
RAP1B
|
RAP1B, member of RAS oncogene family
|
|
chr6_+_43543877
|
0.107
|
NM_006502
|
POLH
|
polymerase (DNA directed), eta
|
|
chr9_+_112542496
|
0.105
|
NM_053016
NM_007203
NM_147150
|
PALM2
PALM2-AKAP2
|
paralemmin 2
PALM2-AKAP2 readthrough
|
|
chr14_-_24020766
|
0.104
|
NM_033400
|
ZFHX2
|
zinc finger homeobox 2
|
|
chr8_-_116681103
|
0.104
|
NM_014112
|
TRPS1
|
trichorhinophalangeal syndrome I
|
|
chr6_+_138483024
|
0.104
|
NM_020340
|
KIAA1244
|
KIAA1244
|
|
chrX_-_103087106
|
0.103
|
NM_016370
|
RAB9B
|
RAB9B, member RAS oncogene family
|
|
chr7_+_28338939
|
0.103
|
NM_182899
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr3_+_196594708
|
0.102
|
NM_152699
|
SENP5
|
SUMO1/sentrin specific peptidase 5
|
|
chr4_-_83933961
|
0.102
|
NM_001115007
|
LIN54
|
lin-54 homolog (C. elegans)
|
|
chr6_+_163835668
|
0.102
|
NM_006775
NM_206853
NM_206854
NM_206855
|
QKI
|
QKI, KH domain containing, RNA binding
|
|
chr15_-_55581969
|
0.100
|
NM_183235
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr4_-_69215699
|
0.100
|
NM_001031732
NM_133370
|
YTHDC1
|
YTH domain containing 1
|
|
chr18_+_55102777
|
0.099
|
NM_004852
|
ONECUT2
|
one cut homeobox 2
|
|
chr17_+_29421941
|
0.098
|
NM_000267
NM_001042492
NM_001128147
|
NF1
|
neurofibromin 1
|
|
chr2_+_139259349
|
0.098
|
NM_001001664
|
SPOPL
|
speckle-type POZ protein-like
|
|
chr8_+_99130067
|
0.098
|
NM_015029
|
POP1
|
processing of precursor 1, ribonuclease P/MRP subunit (S. cerevisiae)
|
|
chr7_-_111846384
|
0.097
|
NM_014705
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr14_-_99737821
|
0.097
|
NM_022898
NM_138576
|
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein)
|
|
chr7_+_28475233
|
0.097
|
NM_004904
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr2_-_172291227
|
0.096
|
NM_024770
|
METTL8
|
methyltransferase like 8
|
|
chr12_-_31479120
|
0.096
|
NM_001135811
NM_021238
NM_001135812
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr5_-_171433876
|
0.094
|
NM_012300
NM_033644
NM_033645
|
FBXW11
|
F-box and WD repeat domain containing 11
|
|
chr1_+_218458628
|
0.094
|
NM_016052
|
RRP15
|
ribosomal RNA processing 15 homolog (S. cerevisiae)
|
|
chr8_+_90769974
|
0.094
|
NM_003821
|
RIPK2
|
receptor-interacting serine-threonine kinase 2
|
|
chr6_-_119031230
|
0.092
|
NM_001178035
|
CEP85L
|
centrosomal protein 85kDa-like
|
|
chr14_+_32546445
|
0.092
|
NM_001030055
NM_001173
|
ARHGAP5
|
Rho GTPase activating protein 5
|
|
chrX_+_49832214
|
0.092
|
NM_000084
|
CLCN5
|
chloride channel 5
|
|
chr5_+_149109814
|
0.091
|
NM_001172698
NM_133263
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr8_+_99129488
|
0.091
|
NM_001145860
NM_001145861
|
POP1
|
processing of precursor 1, ribonuclease P/MRP subunit (S. cerevisiae)
|
|
chr17_+_72733350
|
0.090
|
NM_001006638
NM_001163990
|
RAB37
|
RAB37, member RAS oncogene family
|
|
chr7_+_134576150
|
0.090
|
NM_033139
NM_033140
|
CALD1
|
caldesmon 1
|
|
chr17_+_72732958
|
0.088
|
NM_001163989
|
RAB37
|
RAB37, member RAS oncogene family
|
|
chr6_-_57086204
|
0.087
|
NM_183227
|
RAB23
|
RAB23, member RAS oncogene family
|
|
chr9_+_110045516
|
0.086
|
NM_002874
|
RAD23B
|
RAD23 homolog B (S. cerevisiae)
|
|
chr2_+_70142152
|
0.086
|
NM_001202513
NM_001202514
NM_002357
|
MXD1
|
MAX dimerization protein 1
|
|
chr14_+_32798478
|
0.086
|
NM_004274
|
AKAP6
|
A kinase (PRKA) anchor protein 6
|
|
chr3_-_24536176
|
0.085
|
NM_000461
NM_001128176
NM_001128177
NM_001252634
|
THRB
|
thyroid hormone receptor, beta
|
|
chr1_+_167691156
|
0.083
|
NM_001146191
NM_003953
NM_024569
|
MPZL1
|
myelin protein zero-like 1
|
|
chr1_+_205538111
|
0.083
|
NM_181644
|
MFSD4
|
major facilitator superfamily domain containing 4
|
|
chr17_-_36105005
|
0.082
|
NM_000458
NM_001165923
|
HNF1B
|
HNF1 homeobox B
|
|
chr12_-_25403824
|
0.082
|
NM_004985
NM_033360
|
KRAS
|
v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog
|
|
chr1_+_26438267
|
0.080
|
NM_001243533
NM_152835
|
PDIK1L
|
PDLIM1 interacting kinase 1 like
|
|
chr6_+_37787266
|
0.080
|
NM_021943
|
ZFAND3
|
zinc finger, AN1-type domain 3
|
|
chr11_-_86666211
|
0.079
|
NM_012193
|
FZD4
|
frizzled family receptor 4
|
|
chr4_+_39699663
|
0.079
|
NM_001111112
NM_001111113
NM_005339
|
UBE2K
|
ubiquitin-conjugating enzyme E2K
|
|
chr9_-_23826062
|
0.079
|
NM_004432
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr5_+_176730812
|
0.079
|
NM_013237
|
PRELID1
|
PRELI domain containing 1
|
|
chrX_+_11129405
|
0.079
|
NM_001122608
NM_001171991
NM_005333
|
HCCS
|
holocytochrome c synthase
|
|
chr14_+_29236171
|
0.079
|
NM_005249
|
FOXG1
|
forkhead box G1
|
|
chr8_-_82598517
|
0.078
|
NM_001144878
NM_001144879
NM_005536
|
IMPA1
|
inositol(myo)-1(or 4)-monophosphatase 1
|
|
chr6_-_57087018
|
0.076
|
NM_016277
|
RAB23
|
RAB23, member RAS oncogene family
|
|
chr6_+_64281910
|
0.076
|
NM_003463
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr2_+_110371887
|
0.075
|
NM_023016
|
ANKRD57
|
ankyrin repeat domain 57
|
|
chr20_-_48530275
|
0.075
|
NM_001135773
|
SPATA2
|
spermatogenesis associated 2
|
|
chr3_+_50712671
|
0.075
|
NM_004947
|
DOCK3
|
dedicator of cytokinesis 3
|
|
chr11_-_75062650
|
0.075
|
NM_004041
NM_020251
|
ARRB1
|
arrestin, beta 1
|
|
chr4_-_159592979
|
0.075
|
NM_001008393
|
C4orf46
|
chromosome 4 open reading frame 46
|
|
chr12_-_93323088
|
0.073
|
NM_003566
|
EEA1
|
early endosome antigen 1
|
|
chr11_-_85780899
|
0.072
|
NM_001206947
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr8_+_76452202
|
0.072
|
NM_004133
|
HNF4G
|
hepatocyte nuclear factor 4, gamma
|
|
chr4_-_76598611
|
0.071
|
NM_203504
NM_203505
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2
|
|
chr4_+_40058507
|
0.071
|
NM_018177
|
N4BP2
|
NEDD4 binding protein 2
|
|
chr3_+_147127151
|
0.070
|
NM_003412
|
ZIC1
|
Zic family member 1
|
|
chr1_+_203444882
|
0.070
|
NM_002725
NM_201348
|
PRELP
|
proline/arginine-rich end leucine-rich repeat protein
|
|
chr4_-_76598289
|
0.069
|
NM_012297
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2
|
|
chr8_+_92082423
|
0.069
|
NM_016023
|
OTUD6B
|
OTU domain containing 6B
|
|
chr20_-_48532067
|
0.069
|
NM_006038
|
SPATA2
|
spermatogenesis associated 2
|
|
chr12_-_109251333
|
0.069
|
NM_001161330
NM_018984
|
SSH1
|
slingshot homolog 1 (Drosophila)
|
|
chr10_-_118765087
|
0.069
|
NM_001127211
NM_018330
|
KIAA1598
|
KIAA1598
|
|
chr6_+_126102278
|
0.068
|
NM_001199619
NM_001199620
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr4_-_83931973
|
0.068
|
NM_001115008
NM_194282
|
LIN54
|
lin-54 homolog (C. elegans)
|
|
chr11_+_118477212
|
0.068
|
NM_015157
|
PHLDB1
|
pleckstrin homology-like domain, family B, member 1
|
|
chr4_+_72052354
|
0.068
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr4_-_146100793
|
0.067
|
NM_001102653
|
OTUD4
|
OTU domain containing 4
|
|
chr10_+_86088156
|
0.067
|
NM_018999
|
FAM190B
|
family with sequence similarity 190, member B
|
|
chr12_+_99038918
|
0.067
|
NM_001160
NM_013229
NM_181861
NM_181868
NM_181869
|
APAF1
|
apoptotic peptidase activating factor 1
|
|
chr16_-_9057250
|
0.066
|
NM_003470
|
USP7
|
ubiquitin specific peptidase 7 (herpes virus-associated)
|
|
chr9_-_23821477
|
0.065
|
NM_001171197
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr11_+_118478305
|
0.065
|
NM_001144758
NM_001144759
|
PHLDB1
|
pleckstrin homology-like domain, family B, member 1
|
|
chr20_+_43514340
|
0.065
|
NM_003404
NM_139323
|
YWHAB
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide
|
|
chr5_+_71403040
|
0.065
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr5_-_60458228
|
0.065
|
NM_001048249
|
C5orf43
|
chromosome 5 open reading frame 43
|
|
chr3_-_16306283
|
0.065
|
NM_001047434
NM_206831
|
DPH3
|
DPH3, KTI11 homolog (S. cerevisiae)
|
|
chr16_+_7382750
|
0.064
|
NM_145891
NM_145892
NM_145893
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr5_+_135468522
|
0.062
|
NM_001001419
NM_001001420
NM_005903
|
SMAD5
|
SMAD family member 5
|
|
chr10_+_25464289
|
0.062
|
NM_020752
|
GPR158
|
G protein-coupled receptor 158
|
|
chr9_-_23821842
|
0.062
|
NM_001171195
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chrX_-_129402753
|
0.061
|
NM_017666
|
ZNF280C
|
zinc finger protein 280C
|
|
chr2_-_95825262
|
0.061
|
NM_032788
|
ZNF514
|
zinc finger protein 514
|
|
chr15_-_62352565
|
0.060
|
NM_001018088
NM_017684
NM_018080
NM_020821
|
VPS13C
|
vacuolar protein sorting 13 homolog C (S. cerevisiae)
|
|
chr4_+_57774035
|
0.059
|
NM_005612
|
REST
|
RE1-silencing transcription factor
|
|
chr2_-_60780404
|
0.059
|
NM_018014
NM_022893
NM_138559
|
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein)
|
|
chr9_+_33025207
|
0.058
|
NM_001539
|
DNAJA1
|
DnaJ (Hsp40) homolog, subfamily A, member 1
|
|
chr5_+_149151502
|
0.058
|
NM_001172699
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr10_-_7453412
|
0.057
|
NM_001029880
|
SFMBT2
|
Scm-like with four mbt domains 2
|
|
chr2_+_166428845
|
0.057
|
NM_024969
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr6_-_13814546
|
0.057
|
NM_001031713
|
CCDC90A
|
coiled-coil domain containing 90A
|
|
chr7_+_7222142
|
0.057
|
NM_020156
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1
|
|
chr14_+_100705101
|
0.056
|
NM_003403
|
YY1
|
YY1 transcription factor
|
|
chr2_-_202507348
|
0.056
|
NM_152388
|
TMEM237
|
transmembrane protein 237
|
|
chr21_-_37432789
|
0.056
|
NM_017438
|
SETD4
|
SET domain containing 4
|
|
chr9_+_115983807
|
0.056
|
NM_001859
|
SLC31A1
|
solute carrier family 31 (copper transporters), member 1
|
|
chr8_-_30706532
|
0.055
|
NM_031271
|
TEX15
|
testis expressed 15
|
|
chrX_+_152599544
|
0.055
|
NM_001080485
|
ZNF275
|
zinc finger protein 275
|
|
chr1_-_212004113
|
0.055
|
NM_014873
|
LPGAT1
|
lysophosphatidylglycerol acyltransferase 1
|
|
chr4_+_57775064
|
0.055
|
NM_001193508
|
REST
|
RE1-silencing transcription factor
|
|
chr7_+_91875411
|
0.055
|
NM_019004
|
ANKIB1
|
ankyrin repeat and IBR domain containing 1
|
|
chr1_-_146644045
|
0.054
|
NM_005399
|
PRKAB2
|
protein kinase, AMP-activated, beta 2 non-catalytic subunit
|
|
chr6_-_7313352
|
0.054
|
NM_003144
|
SSR1
|
signal sequence receptor, alpha
|
|
chr16_-_12009804
|
0.054
|
NM_001130006
NM_002094
|
GSPT1
|
G1 to S phase transition 1
|
|
chr12_-_48499415
|
0.054
|
NM_014554
|
SENP1
|
SUMO1/sentrin specific peptidase 1
|
|
chr12_+_65218351
|
0.054
|
NM_015279
|
TBC1D30
|
TBC1 domain family, member 30
|
|
chr18_+_67068134
|
0.053
|
NM_152721
|
DOK6
|
docking protein 6
|
|
chr16_-_12010518
|
0.052
|
NM_001130007
|
GSPT1
|
G1 to S phase transition 1
|
|
chr4_-_23891608
|
0.051
|
NM_013261
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha
|