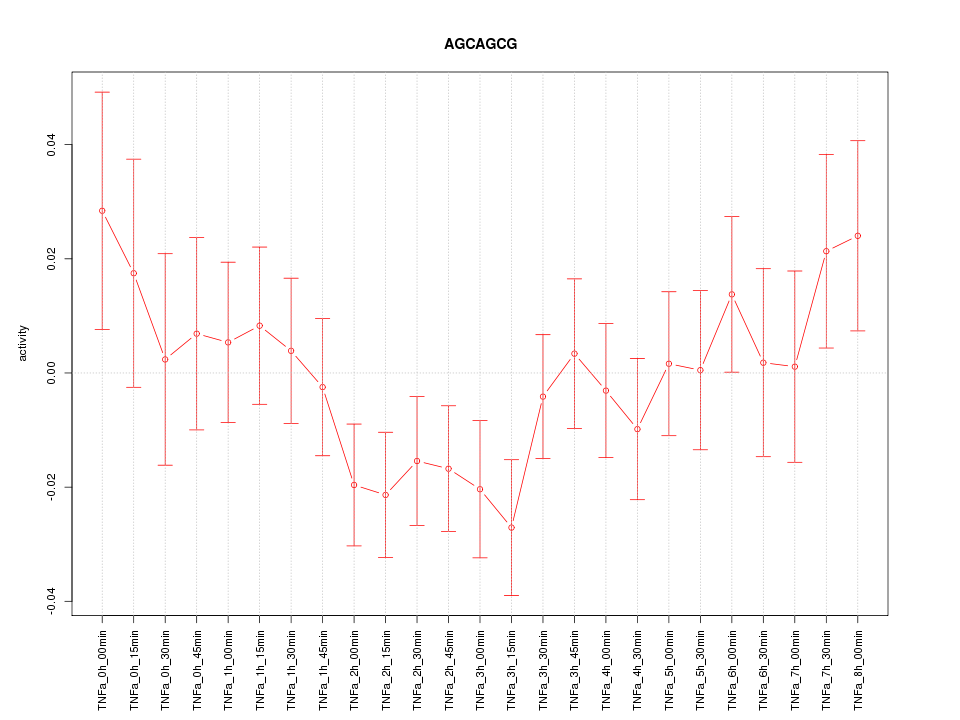
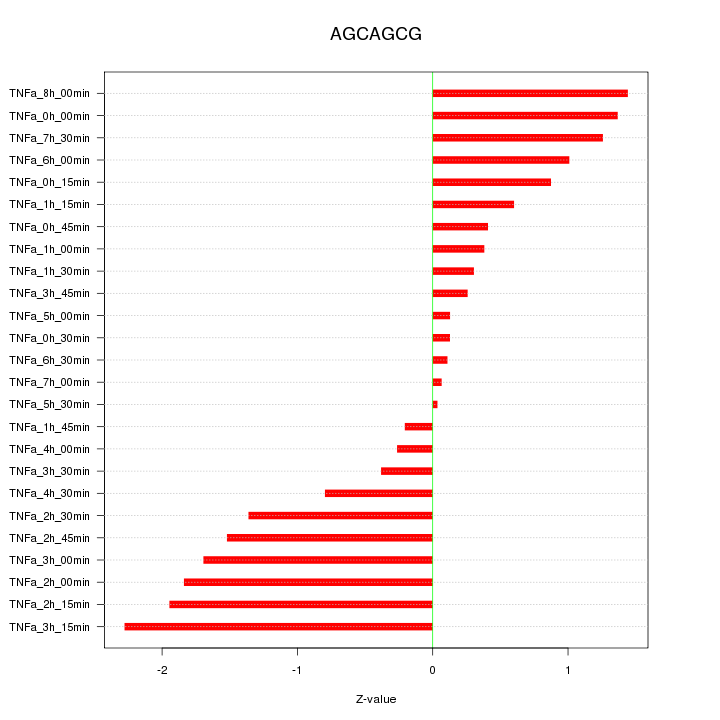
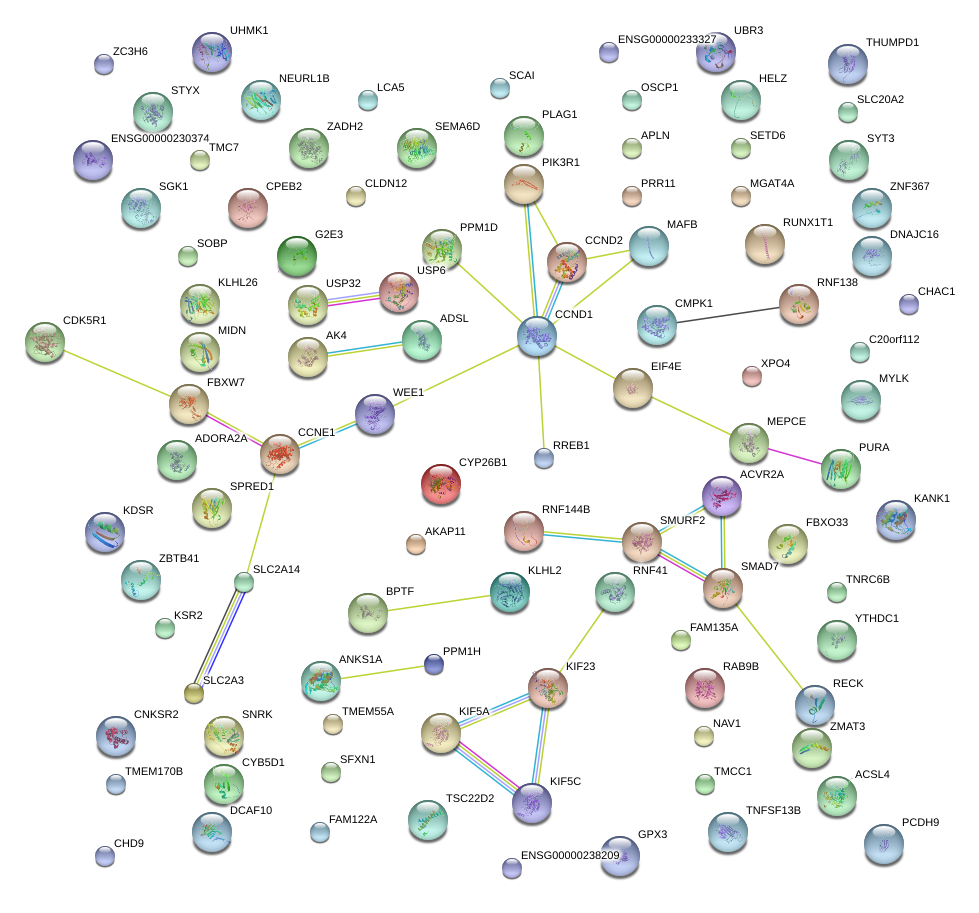
|
chr18_-_46477080
|
2.316
|
NM_001190821
NM_005904
|
SMAD7
|
SMAD family member 7
|
|
chr18_-_46475702
|
2.278
|
NM_001190822
|
SMAD7
|
SMAD family member 7
|
|
chr18_-_46469118
|
2.175
|
NM_001190823
|
SMAD7
|
SMAD family member 7
|
|
chr6_+_7107986
|
1.865
|
NM_001003698
NM_001003699
NM_001003700
|
RREB1
|
ras responsive element binding protein 1
|
|
chr9_+_504661
|
1.853
|
NM_015158
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr9_+_706744
|
1.795
|
NM_153186
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr6_+_7107799
|
1.754
|
NM_001168344
|
RREB1
|
ras responsive element binding protein 1
|
|
chr3_+_43328001
|
1.626
|
NM_001100594
NM_017719
|
SNRK
|
SNF related kinase
|
|
chr11_+_69455837
|
1.586
|
NM_053056
|
CCND1
|
cyclin D1
|
|
chr8_-_57123858
|
1.442
|
NM_001114634
NM_001114635
NM_002655
|
PLAG1
|
pleiomorphic adenoma gene 1
|
|
chr8_-_93115453
|
1.370
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chrX_-_108976509
|
1.202
|
NM_004458
NM_022977
|
ACSL4
|
acyl-CoA synthetase long-chain family member 4
|
|
chr8_-_93107881
|
1.199
|
NM_001198625
NM_001198626
NM_001198627
NM_001198631
NM_001198634
NM_001198679
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_-_93111915
|
1.187
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_-_93107442
|
1.184
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_-_93029864
|
1.159
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr17_+_58677489
|
1.156
|
NM_003620
|
PPM1D
|
protein phosphatase, Mg2+/Mn2+ dependent, 1D
|
|
chr6_+_18387580
|
1.151
|
NM_182757
|
RNF144B
|
ring finger protein 144B
|
|
chr12_-_8088629
|
1.123
|
NM_006931
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3
|
|
chr8_-_93075190
|
1.113
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr19_-_51143091
|
1.096
|
NM_001160328
NM_001160329
|
SYT3
|
synaptotagmin III
|
|
chr19_-_51141301
|
1.016
|
NM_032298
|
SYT3
|
synaptotagmin III
|
|
chr20_-_31071187
|
1.012
|
NM_080616
|
C20orf112
|
chromosome 20 open reading frame 112
|
|
chr22_+_40440664
|
0.990
|
NM_001024843
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr3_-_129612371
|
0.965
|
NM_001128224
|
TMCC1
|
transmembrane and coiled-coil domain family 1
|
|
chr8_-_92053043
|
0.951
|
NM_018710
|
TMEM55A
|
transmembrane protein 55A
|
|
chr13_-_67804432
|
0.945
|
NM_020403
NM_203487
|
PCDH9
|
protocadherin 9
|
|
chr16_-_20753117
|
0.928
|
NM_017736
|
THUMPD1
|
THUMP domain containing 1
|
|
chr18_-_72921280
|
0.926
|
NM_175907
|
ZADH2
|
zinc binding alcohol dehydrogenase domain containing 2
|
|
chr3_-_123603144
|
0.926
|
NM_053025
NM_053026
NM_053027
NM_053028
|
MYLK
|
myosin light chain kinase
|
|
chr6_-_134497069
|
0.923
|
NM_001143678
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr5_+_67511578
|
0.918
|
NM_181523
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr1_+_65613211
|
0.913
|
NM_203464
NM_001005353
|
AK4
|
adenylate kinase 4
|
|
chr1_-_197169635
|
0.907
|
NM_194314
|
ZBTB41
|
zinc finger and BTB domain containing 41
|
|
chr15_+_47476225
|
0.907
|
NM_001198999
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr20_-_39317875
|
0.904
|
NM_005461
|
MAFB
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian)
|
|
chr1_+_15853348
|
0.885
|
NM_015291
|
DNAJC16
|
DnaJ (Hsp40) homolog, subfamily C, member 16
|
|
chr6_-_134498973
|
0.876
|
NM_001143677
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr3_-_123339423
|
0.875
|
NM_053031
NM_053032
|
MYLK
|
myosin light chain kinase
|
|
chr16_+_53088791
|
0.872
|
NM_025134
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr13_+_42846255
|
0.858
|
NM_016248
|
AKAP11
|
A kinase (PRKA) anchor protein 11
|
|
chr1_+_65613771
|
0.856
|
NM_013410
|
AK4
|
adenylate kinase 4
|
|
chr6_-_134496006
|
0.855
|
NM_005627
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr6_+_34857021
|
0.845
|
NM_015245
|
ANKS1A
|
ankyrin repeat and sterile alpha motif domain containing 1A
|
|
chr5_+_174905513
|
0.831
|
NM_022754
|
SFXN1
|
sideroflexin 1
|
|
chr19_+_30302861
|
0.829
|
NM_001238
|
CCNE1
|
cyclin E1
|
|
chrX_-_128788913
|
0.819
|
NM_017413
|
APLN
|
apelin
|
|
chr22_+_24823529
|
0.814
|
NM_000675
|
ADORA2A
|
adenosine A2a receptor
|
|
chr6_+_107811272
|
0.806
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr6_-_134639195
|
0.806
|
NM_001143676
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr3_-_129599220
|
0.802
|
NM_001017395
|
TMCC1
|
transmembrane and coiled-coil domain family 1
|
|
chr12_-_56615484
|
0.798
|
NM_001242826
|
RNF41
|
ring finger protein 41
|
|
chr6_+_71123105
|
0.796
|
NM_001105531
NM_020819
|
FAM135A
|
family with sequence similarity 135, member A
|
|
chr5_+_67586466
|
0.768
|
NM_181504
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr6_+_11538486
|
0.768
|
NM_001100829
|
TMEM170B
|
transmembrane protein 170B
|
|
chr2_+_113033146
|
0.751
|
NM_198581
|
ZC3H6
|
zinc finger CCCH-type containing 6
|
|
chr5_+_139493689
|
0.734
|
NM_005859
|
PURA
|
purine-rich element binding protein A
|
|
chr15_+_48010685
|
0.734
|
NM_020858
NM_024966
NM_153616
NM_153617
NM_153618
NM_153619
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr5_+_67584251
|
0.720
|
NM_181524
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr12_-_56615679
|
0.713
|
NM_005785
NM_194358
NM_194359
|
RNF41
|
ring finger protein 41
|
|
chr1_+_162466963
|
0.711
|
NM_001184763
|
UHMK1
|
U2AF homology motif (UHM) kinase 1
|
|
chr6_+_71136141
|
0.696
|
NM_001162529
|
FAM135A
|
family with sequence similarity 135, member A
|
|
chr1_+_162467594
|
0.692
|
NM_144624
NM_175866
|
UHMK1
|
U2AF homology motif (UHM) kinase 1
|
|
chr13_+_108921976
|
0.687
|
NM_001145645
NM_006573
|
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b
|
|
chr11_+_9596233
|
0.686
|
NM_001143976
|
WEE1
|
WEE1 homolog (S. pombe)
|
|
chr4_-_69215699
|
0.678
|
NM_001031732
NM_133370
|
YTHDC1
|
YTH domain containing 1
|
|
chr17_-_62658311
|
0.677
|
NM_022739
|
SMURF2
|
SMAD specific E3 ubiquitin protein ligase 2
|
|
chr17_+_30813928
|
0.662
|
NM_003885
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35)
|
|
chr17_+_7761063
|
0.652
|
NM_144607
|
CYB5D1
|
cytochrome b5 domain containing 1
|
|
chr3_-_178789597
|
0.643
|
NM_022470
NM_152240
|
ZMAT3
|
zinc finger, matrin-type 3
|
|
chr9_+_71394919
|
0.610
|
NM_138333
|
FAM122A
|
family with sequence similarity 122A
|
|
chr5_+_67588395
|
0.608
|
NM_001242466
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr22_+_40573879
|
0.604
|
NM_001162501
NM_015088
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr12_+_57943843
|
0.601
|
NM_004984
|
KIF5A
|
kinesin family member 5A
|
|
chr9_-_99180661
|
0.600
|
NM_153695
|
ZNF367
|
zinc finger protein 367
|
|
chr17_-_65241305
|
0.598
|
NM_014877
|
HELZ
|
helicase with zinc finger
|
|
chr14_+_53196852
|
0.596
|
NM_001130701
NM_145251
|
STYX
|
serine/threonine/tyrosine interacting protein
|
|
chr7_+_90032647
|
0.591
|
NM_001185072
NM_001185073
NM_012129
|
CLDN12
|
claudin 12
|
|
chr12_-_118406027
|
0.584
|
NM_173598
|
KSR2
|
kinase suppressor of ras 2
|
|
chr9_+_37800779
|
0.577
|
NM_024345
|
DCAF10
|
DDB1 and CUL4 associated factor 10
|
|
chr17_+_65821625
|
0.574
|
NM_004459
NM_182641
|
BPTF
|
bromodomain PHD finger transcription factor
|
|
chr1_+_201617449
|
0.573
|
NM_020443
|
NAV1
|
neuron navigator 1
|
|
chr2_+_148602569
|
0.569
|
NM_001616
|
ACVR2A
|
activin A receptor, type IIA
|
|
chr19_+_1248548
|
0.565
|
NM_177401
|
MIDN
|
midnolin
|
|
chr4_+_166128769
|
0.564
|
NM_007246
|
KLHL2
|
kelch-like 2, Mayven (Drosophila)
|
|
chr9_-_127905715
|
0.564
|
NM_001144877
NM_173690
|
SCAI
|
suppressor of cancer cell invasion
|
|
chr4_+_166131170
|
0.561
|
NM_001161521
NM_001161522
|
KLHL2
|
kelch-like 2, Mayven (Drosophila)
|
|
chr12_+_4382882
|
0.554
|
NM_001759
|
CCND2
|
cyclin D2
|
|
chr15_+_38544966
|
0.551
|
NM_152594
|
SPRED1
|
sprouty-related, EVH1 domain containing 1
|
|
chr19_+_18747827
|
0.550
|
NM_018316
|
KLHL26
|
kelch-like 26 (Drosophila)
|
|
chr14_-_39901619
|
0.548
|
NM_203301
|
FBXO33
|
F-box protein 33
|
|
chr4_-_99851785
|
0.544
|
NM_001130679
NM_001968
|
EIF4E
|
eukaryotic translation initiation factor 4E
|
|
chr16_+_18995508
|
0.537
|
NM_001160364
|
TMC7
|
transmembrane channel-like 7
|
|
chr11_+_9595183
|
0.534
|
NM_003390
|
WEE1
|
WEE1 homolog (S. pombe)
|
|
chr18_-_61034350
|
0.531
|
NM_002035
|
KDSR
|
3-ketodihydrosphingosine reductase
|
|
chrX_-_103087106
|
0.530
|
NM_016370
|
RAB9B
|
RAB9B, member RAS oncogene family
|
|
chr3_+_150126705
|
0.510
|
NM_014779
|
TSC22D2
|
TSC22 domain family, member 2
|
|
chr8_-_42397048
|
0.507
|
NM_006749
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2
|
|
chr1_-_36915969
|
0.500
|
NM_145047
NM_206837
|
OSCP1
|
organic solute carrier partner 1
|
|
chr1_+_201708940
|
0.498
|
NM_001167738
|
NAV1
|
neuron navigator 1
|
|
chr4_+_15004297
|
0.478
|
NM_001177381
NM_001177382
NM_001177383
NM_001177384
NM_182485
NM_182646
|
CPEB2
|
cytoplasmic polyadenylation element binding protein 2
|
|
chr16_+_18995255
|
0.471
|
NM_024847
|
TMC7
|
transmembrane channel-like 7
|
|
chr14_+_31028328
|
0.469
|
NM_017769
|
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase
|
|
chr5_+_172068188
|
0.464
|
NM_001142651
|
NEURL1B
|
neuralized homolog 1B (Drosophila)
|
|
chr4_-_99850242
|
0.461
|
NM_001130678
|
EIF4E
|
eukaryotic translation initiation factor 4E
|
|
chr1_+_47799447
|
0.460
|
NM_001136140
NM_016308
|
CMPK1
|
cytidine monophosphate (UMP-CMP) kinase 1, cytosolic
|
|
chr6_-_80247103
|
0.460
|
NM_001122769
NM_181714
|
LCA5
|
Leber congenital amaurosis 5
|
|
chr18_+_29672568
|
0.459
|
NM_001191324
NM_198128
|
RNF138
|
ring finger protein 138
|
|
chr4_-_153456152
|
0.439
|
NM_033632
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr12_-_63328663
|
0.435
|
NM_020700
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr16_+_58549382
|
0.435
|
NM_001160305
NM_024860
|
SETD6
|
SET domain containing 6
|
|
chr17_+_5031686
|
0.434
|
NM_004505
|
USP6
|
ubiquitin specific peptidase 6 (Tre-2 oncogene)
|
|
chr2_-_99347588
|
0.433
|
NM_012214
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chr13_-_21476895
|
0.429
|
NM_022459
|
XPO4
|
exportin 4
|
|
chr9_+_36036900
|
0.428
|
NM_021111
|
RECK
|
reversion-inducing-cysteine-rich protein with kazal motifs
|
|
chr18_+_29671817
|
0.428
|
NM_016271
|
RNF138
|
ring finger protein 138
|
|
chr15_+_69706619
|
0.428
|
NM_004856
NM_138555
|
KIF23
|
kinesin family member 23
|
|
chr4_-_153303663
|
0.425
|
NM_001013415
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr2_+_170683900
|
0.415
|
NM_172070
|
UBR3
|
ubiquitin protein ligase E3 component n-recognin 3 (putative)
|
|
chr15_+_41245635
|
0.415
|
NM_001142776
NM_024111
|
CHAC1
|
ChaC, cation transport regulator homolog 1 (E. coli)
|
|
chr12_-_8025419
|
0.411
|
NM_153449
|
SLC2A14
|
solute carrier family 2 (facilitated glucose transporter), member 14
|
|
chr2_-_72374919
|
0.409
|
NM_019885
|
CYP26B1
|
cytochrome P450, family 26, subfamily B, polypeptide 1
|
|
chr7_+_100026412
|
0.405
|
NM_001194990
NM_001194991
NM_001194992
|
MEPCE
|
methylphosphate capping enzyme
|
|
chr17_+_57232859
|
0.405
|
NM_018304
|
PRR11
|
proline rich 11
|
|
chr2_-_99279935
|
0.404
|
NM_001160154
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chr3_+_37903643
|
0.382
|
NM_001008392
NM_005808
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like
|
|
chr21_+_17102370
|
0.376
|
NM_013396
|
USP25
|
ubiquitin specific peptidase 25
|
|
chr10_-_94003002
|
0.376
|
NM_001178137
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr7_+_100027253
|
0.375
|
NM_019606
|
MEPCE
|
methylphosphate capping enzyme
|
|
chr13_+_20532788
|
0.371
|
NM_003453
NM_197968
NM_001190964
|
ZMYM2
|
zinc finger, MYM-type 2
|
|
chr11_+_111848008
|
0.371
|
NM_033425
|
DIXDC1
|
DIX domain containing 1
|
|
chr5_+_140186658
|
0.370
|
NM_018907
NM_031500
|
PCDHA4
|
protocadherin alpha 4
|
|
chr5_+_140213968
|
0.370
|
NM_018910
NM_031852
|
PCDHA7
|
protocadherin alpha 7
|
|
chr4_-_153273683
|
0.367
|
NM_018315
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr6_-_144385647
|
0.364
|
NM_001080951
NM_001080955
|
PLAGL1
|
pleiomorphic adenoma gene-like 1
|
|
chr11_-_110583120
|
0.364
|
NM_020809
|
ARHGAP20
|
Rho GTPase activating protein 20
|
|
chr8_+_21777165
|
0.362
|
NM_015024
|
XPO7
|
exportin 7
|
|
chr1_-_155532322
|
0.356
|
NM_018489
|
ASH1L
|
ash1 (absent, small, or homeotic)-like (Drosophila)
|
|
chr2_-_171923482
|
0.355
|
NM_001136555
|
TLK1
|
tousled-like kinase 1
|
|
chr9_+_4679557
|
0.355
|
NM_017913
|
CDC37L1
|
cell division cycle 37 homolog (S. cerevisiae)-like 1
|
|
chr5_+_140220811
|
0.355
|
NM_018911
NM_031856
|
PCDHA8
|
protocadherin alpha 8
|
|
chr5_+_140254930
|
0.354
|
NM_018903
NM_031864
|
PCDHA12
|
protocadherin alpha 12
|
|
chr1_-_48462561
|
0.339
|
NM_001194986
|
LOC388630
|
UPF0632 protein A
|
|
chr1_-_22469414
|
0.339
|
NM_030761
|
WNT4
|
wingless-type MMTV integration site family, member 4
|
|
chr1_+_184356128
|
0.338
|
NM_030806
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr6_+_15246299
|
0.338
|
NM_004973
|
JARID2
|
jumonji, AT rich interactive domain 2
|
|
chr22_-_42017032
|
0.330
|
NM_015704
|
PPPDE2
|
PPPDE peptidase domain containing 2
|
|
chr1_+_10092990
|
0.327
|
NM_001105562
NM_006048
|
UBE4B
|
ubiquitination factor E4B
|
|
chr13_-_50367027
|
0.320
|
NM_002267
|
KPNA3
|
karyopherin alpha 3 (importin alpha 4)
|
|
chr19_-_17799005
|
0.309
|
NM_001080421
|
UNC13A
|
unc-13 homolog A (C. elegans)
|
|
chr6_-_144329342
|
0.308
|
NM_001080952
NM_001080953
NM_001080954
NM_001080956
NM_002656
NM_006718
|
PLAGL1
HYMAI
|
pleiomorphic adenoma gene-like 1
hydatidiform mole associated and imprinted (non-protein coding)
|
|
chr3_+_187871473
|
0.306
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr2_+_198381021
|
0.305
|
NM_001204094
|
MOB4
|
MOB family member 4, phocein
|
|
chr5_-_16465856
|
0.305
|
NM_033414
|
ZNF622
|
zinc finger protein 622
|
|
chr17_+_40834571
|
0.304
|
NM_003632
|
CNTNAP1
|
contactin associated protein 1
|
|
chr11_+_9406123
|
0.298
|
NM_006391
|
IPO7
|
importin 7
|
|
chr20_+_44650260
|
0.295
|
NM_001134771
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5
|
|
chr18_+_19321544
|
0.291
|
NM_020774
|
MIB1
|
mindbomb homolog 1 (Drosophila)
|
|
chr11_+_125496123
|
0.291
|
NM_001274
|
CHEK1
|
checkpoint kinase 1
|
|
chr20_-_52199635
|
0.290
|
NM_006526
|
ZNF217
|
zinc finger protein 217
|
|
chr5_+_140180782
|
0.289
|
NM_018906
NM_031497
|
PCDHA3
|
protocadherin alpha 3
|
|
chr2_-_220408264
|
0.288
|
NM_024536
|
CHPF
|
chondroitin polymerizing factor
|
|
chr14_+_102228134
|
0.278
|
NM_001161725
NM_001161726
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr15_-_29862926
|
0.277
|
NM_015307
|
FAM189A1
|
family with sequence similarity 189, member A1
|
|
chr18_+_47088400
|
0.277
|
NM_006033
|
LIPG
|
lipase, endothelial
|
|
chr11_-_130184459
|
0.273
|
NM_014155
|
ZBTB44
|
zinc finger and BTB domain containing 44
|
|
chr8_+_67687415
|
0.270
|
NM_013257
NM_170709
|
SGK3
|
serum/glucocorticoid regulated kinase family, member 3
|
|
chr7_-_15726273
|
0.268
|
NM_005924
|
MEOX2
|
mesenchyme homeobox 2
|
|
chr12_-_112037360
|
0.267
|
NM_002973
|
ATXN2
|
ataxin 2
|
|
chr2_-_172017358
|
0.267
|
NM_012290
|
TLK1
|
tousled-like kinase 1
|
|
chr20_+_19193282
|
0.264
|
NM_020689
|
SLC24A3
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3
|
|
chr16_-_47007567
|
0.263
|
NM_005880
|
DNAJA2
|
DnaJ (Hsp40) homolog, subfamily A, member 2
|
|
chr2_-_172087823
|
0.262
|
NM_001136554
|
TLK1
|
tousled-like kinase 1
|
|
chr10_-_94050799
|
0.261
|
NM_014912
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr9_-_111882209
|
0.259
|
NM_032012
|
C9orf5
|
chromosome 9 open reading frame 5
|
|
chr13_-_36429798
|
0.259
|
NM_001195415
NM_001195416
NM_001195430
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr10_-_79789290
|
0.257
|
NM_007055
|
POLR3A
|
polymerase (RNA) III (DNA directed) polypeptide A, 155kDa
|
|
chr12_-_56881901
|
0.252
|
NM_013267
|
GLS2
|
glutaminase 2 (liver, mitochondrial)
|
|
chr7_+_30323922
|
0.251
|
NM_147128
|
ZNRF2
|
zinc and ring finger 2
|
|
chr20_+_44657812
|
0.251
|
NM_020708
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5
|
|
chr1_+_2160133
|
0.251
|
NM_003036
|
SKI
|
v-ski sarcoma viral oncogene homolog (avian)
|
|
chr3_-_120068077
|
0.250
|
NM_001099678
|
LRRC58
|
leucine rich repeat containing 58
|
|
chr4_+_160188997
|
0.248
|
NM_014247
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2
|
|
chr12_+_66218141
|
0.244
|
NM_003483
NM_003484
|
HMGA2
|
high mobility group AT-hook 2
|
|
chr12_+_83080901
|
0.243
|
NM_152588
|
TMTC2
|
transmembrane and tetratricopeptide repeat containing 2
|
|
chr8_+_67624652
|
0.243
|
NM_001033578
|
SGK3
|
serum/glucocorticoid regulated kinase family, member 3
|
|
chr11_-_65667860
|
0.241
|
NM_005438
|
FOSL1
|
FOS-like antigen 1
|
|
chr1_-_114354906
|
0.240
|
NM_018364
|
RSBN1
|
round spermatid basic protein 1
|
|
chr5_+_140247767
|
0.239
|
NM_018902
NM_031861
|
PCDHA11
|
protocadherin alpha 11
|
|
chr5_+_140201221
|
0.239
|
NM_018908
NM_031501
|
PCDHA5
|
protocadherin alpha 5
|
|
chr2_-_55277698
|
0.237
|
NM_020532
NM_153828
NM_207520
|
RTN4
|
reticulon 4
|
|
chr2_+_198380294
|
0.234
|
NM_199482
|
MOB4
|
MOB family member 4, phocein
|
|
chr16_-_30546145
|
0.233
|
NM_023931
|
ZNF747
|
zinc finger protein 747
|
|
chr9_-_127703294
|
0.230
|
NM_002077
|
GOLGA1
|
golgin A1
|
|
chr1_+_33722158
|
0.229
|
NM_152493
|
ZNF362
|
zinc finger protein 362
|
|
chr10_+_73723992
|
0.227
|
NM_004273
|
CHST3
|
carbohydrate (chondroitin 6) sulfotransferase 3
|
|
chr5_+_140165875
|
0.226
|
NM_018900
NM_031410
NM_031411
|
PCDHA1
|
protocadherin alpha 1
|
|
chr5_+_140207562
|
0.225
|
NM_018909
NM_031848
NM_031849
|
PCDHA6
|
protocadherin alpha 6
|
|
chr17_-_2304217
|
0.225
|
NM_020310
|
MNT
|
MAX binding protein
|
|
chr12_+_122242580
|
0.221
|
NM_015048
|
SETD1B
|
SET domain containing 1B
|