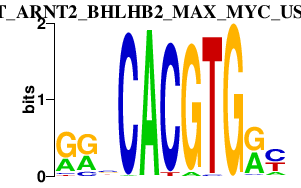
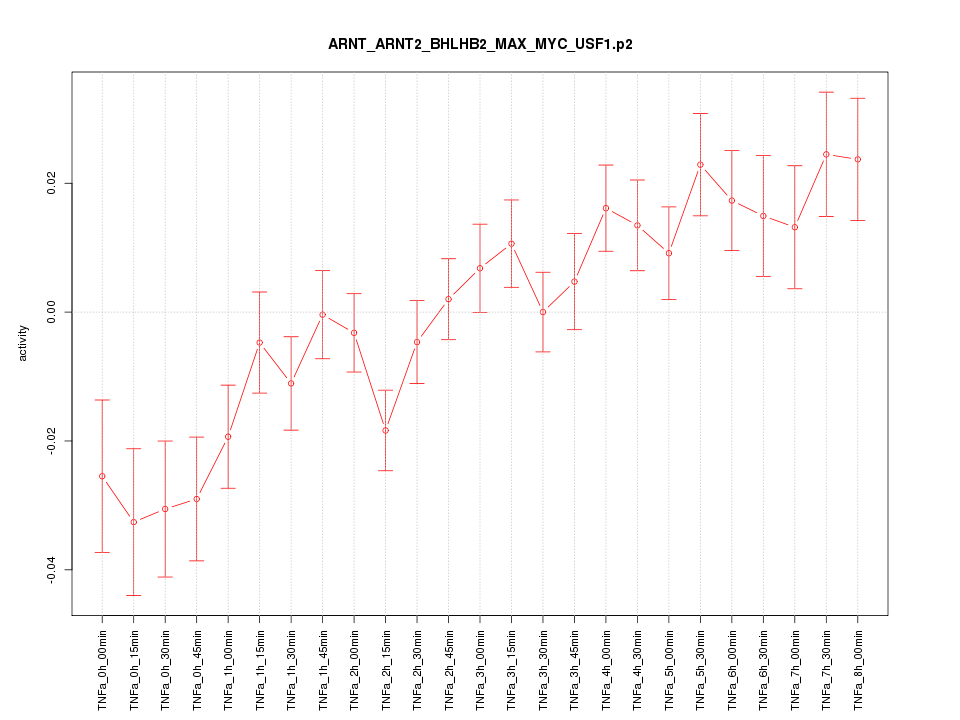
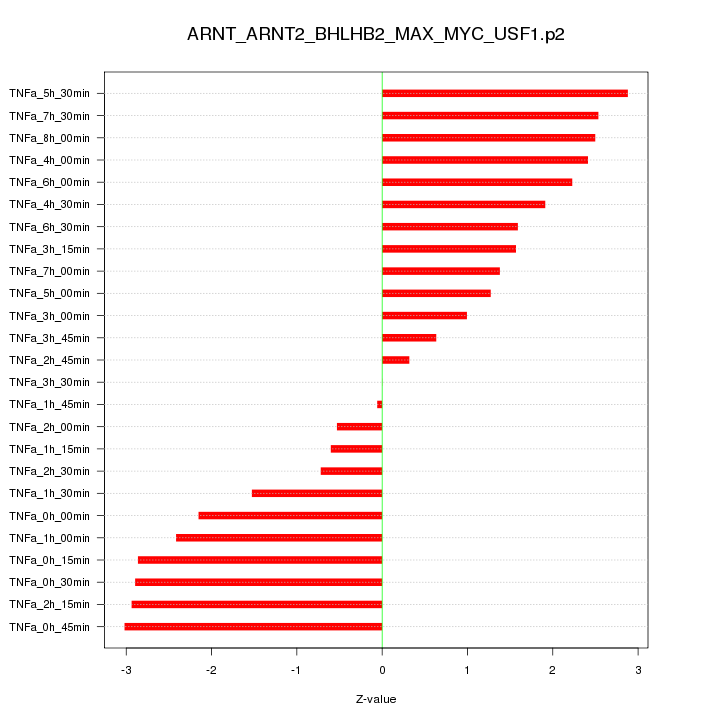
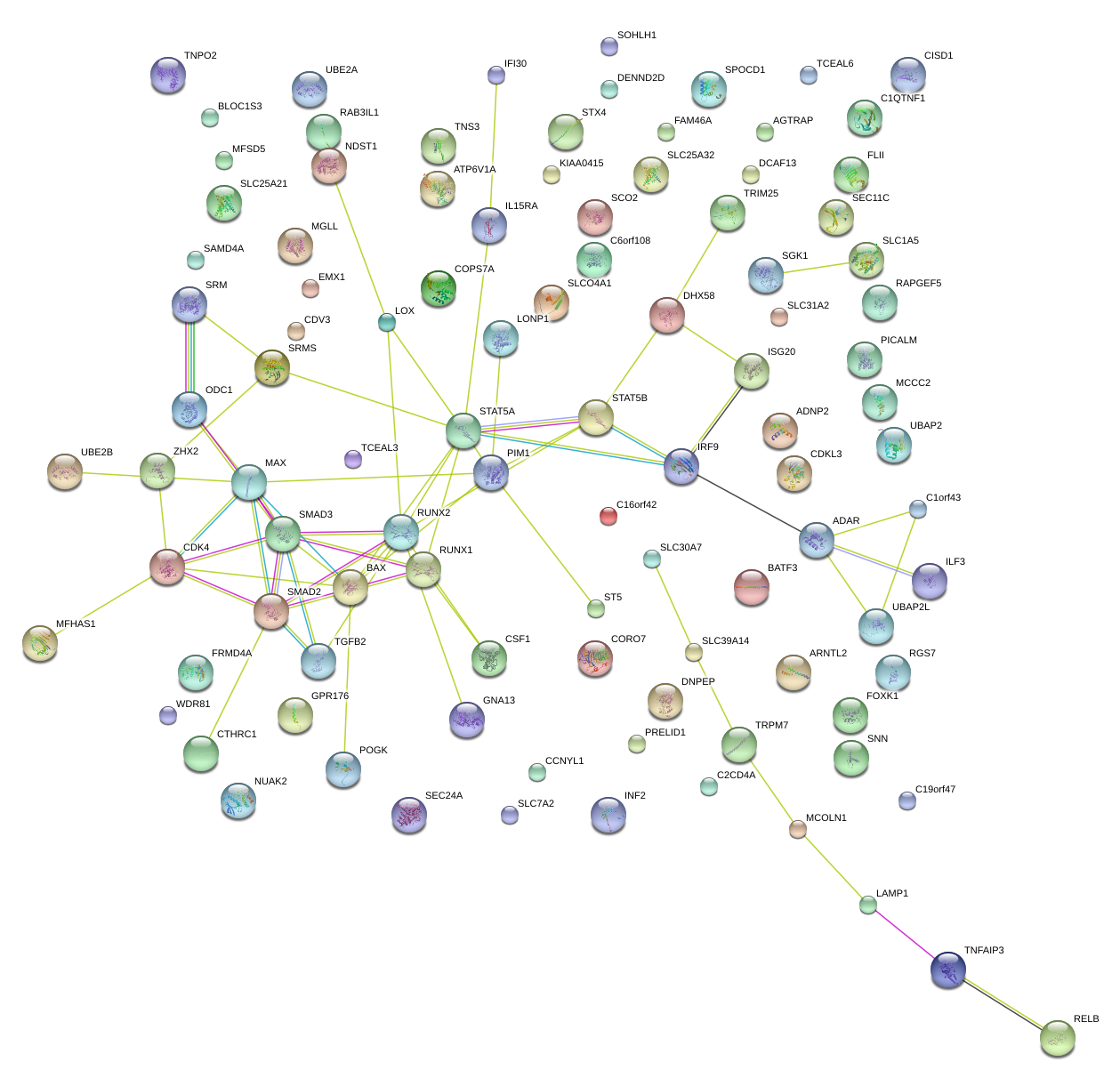
|
chr8_+_17354562
|
16.075
|
NM_001008539
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr15_+_89182038
|
11.500
|
NM_002201
|
ISG20
|
interferon stimulated exonuclease gene 20kDa
|
|
chr19_+_45504686
|
11.496
|
NM_006509
|
RELB
|
v-rel reticuloendotheliosis viral oncogene homolog B
|
|
chr1_-_154193044
|
9.400
|
|
C1orf43
|
chromosome 1 open reading frame 43
|
|
chr9_+_115913227
|
8.304
|
NM_001860
|
SLC31A2
|
solute carrier family 31 (copper transporters), member 2
|
|
chr9_+_115913266
|
8.009
|
|
SLC31A2
|
solute carrier family 31 (copper transporters), member 2
|
|
chr19_+_18284578
|
7.939
|
NM_006332
|
IFI30
|
interferon, gamma-inducible protein 30
|
|
chr7_-_47622127
|
7.857
|
|
TNS3
|
tensin 3
|
|
chr19_+_49458151
|
7.118
|
|
BAX
|
BCL2-associated X protein
|
|
chr1_-_212873306
|
7.081
|
NM_018664
|
BATF3
|
basic leucine zipper transcription factor, ATF-like 3
|
|
chr19_+_49458129
|
6.853
|
|
BAX
|
BCL2-associated X protein
|
|
chr16_+_31044849
|
6.693
|
|
STX4
|
syntaxin 4
|
|
chr19_+_49458031
|
6.550
|
NM_004324
NM_138761
NM_138763
NM_138764
|
BAX
|
BCL2-associated X protein
|
|
chr19_+_10765106
|
6.500
|
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr8_+_104383701
|
6.459
|
|
CTHRC1
|
collagen triple helix repeat containing 1
|
|
chr13_+_113951537
|
6.443
|
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr16_+_31044881
|
6.322
|
NM_004604
|
STX4
|
syntaxin 4
|
|
chr19_+_18284619
|
6.281
|
|
IFI30
|
interferon, gamma-inducible protein 30
|
|
chr19_+_18284631
|
6.220
|
|
IFI30
|
interferon, gamma-inducible protein 30
|
|
chr15_+_67358194
|
6.002
|
NM_005902
|
SMAD3
|
SMAD family member 3
|
|
chr2_-_10588451
|
5.884
|
NM_002539
|
ODC1
|
ornithine decarboxylase 1
|
|
chr14_+_93799564
|
5.882
|
NM_020818
|
UNC79
|
unc-79 homolog (C. elegans)
|
|
chr21_-_36421586
|
5.748
|
NM_001754
|
RUNX1
|
runt-related transcription factor 1
|
|
chr21_-_36421630
|
5.668
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr16_+_31044414
|
5.666
|
|
STX4
|
syntaxin 4
|
|
chr19_+_10764987
|
5.578
|
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr21_+_26934440
|
5.517
|
|
MIR155HG
|
MIR155 host gene (non-protein coding)
|
|
chr13_+_113951461
|
5.449
|
NM_005561
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr11_-_8892909
|
5.340
|
|
ST5
|
suppression of tumorigenicity 5
|
|
chr13_+_113951589
|
5.339
|
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr10_-_6019454
|
5.317
|
NM_002189
NM_172200
|
IL15RA
|
interleukin 15 receptor, alpha
|
|
chr1_+_154193294
|
5.285
|
NM_014847
|
UBAP2L
|
ubiquitin associated protein 2-like
|
|
chr19_+_10764896
|
5.267
|
NM_001137673
NM_004516
NM_012218
NM_017620
NM_153464
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr1_+_11796177
|
5.048
|
|
AGTRAP
|
angiotensin II receptor-associated protein
|
|
chr3_-_127541160
|
4.953
|
NM_007283
|
MGLL
|
monoglyceride lipase
|
|
chr8_+_104383785
|
4.911
|
NM_138455
|
CTHRC1
|
collagen triple helix repeat containing 1
|
|
chr1_+_11796128
|
4.727
|
NM_001040194
NM_001040195
NM_001040196
NM_001040197
NM_020350
|
AGTRAP
|
angiotensin II receptor-associated protein
|
|
chr14_+_24630421
|
4.667
|
NM_006084
|
IRF9
|
interferon regulatory factor 9
|
|
chr19_-_10764449
|
4.595
|
|
LOC147727
|
uncharacterized LOC147727
|
|
chr1_+_11796211
|
4.573
|
|
AGTRAP
|
angiotensin II receptor-associated protein
|
|
chr7_-_47621741
|
4.520
|
NM_022748
|
TNS3
|
tensin 3
|
|
chr1_-_154600446
|
4.426
|
NM_001025107
|
ADAR
|
adenosine deaminase, RNA-specific
|
|
chr1_-_154600383
|
4.389
|
|
ADAR
|
adenosine deaminase, RNA-specific
|
|
chr1_+_101361649
|
4.315
|
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7
|
|
chr1_+_218518675
|
4.303
|
NM_001135599
NM_003238
|
TGFB2
|
transforming growth factor, beta 2
|
|
chr3_-_127541975
|
4.267
|
NM_001003794
|
MGLL
|
monoglyceride lipase
|
|
chr9_-_138591318
|
4.236
|
NM_001012415
NM_001101677
|
SOHLH1
|
spermatogenesis and oogenesis specific basic helix-loop-helix 1
|
|
chr8_-_104427373
|
4.168
|
NM_030780
|
SLC25A32
|
solute carrier family 25, member 32
|
|
chr17_+_40440481
|
4.152
|
|
STAT5A
|
signal transducer and activator of transcription 5A
|
|
chr16_+_11762235
|
4.099
|
NM_003498
|
SNN
|
stannin
|
|
chr6_-_43197178
|
4.071
|
NM_006443
NM_199184
|
C6orf108
|
chromosome 6 open reading frame 108
|
|
chr22_-_50963961
|
4.056
|
NM_001169111
NM_005138
|
SCO2
|
SCO cytochrome oxidase deficient homolog 2 (yeast)
|
|
chr5_+_70883132
|
3.887
|
|
MCCC2
|
methylcrotonoyl-CoA carboxylase 2 (beta)
|
|
chr15_+_67358161
|
3.857
|
|
SMAD3
|
SMAD family member 3
|
|
chr1_-_212873197
|
3.839
|
|
BATF3
|
basic leucine zipper transcription factor, ATF-like 3
|
|
chr17_+_77030471
|
3.711
|
NM_198593
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1
|
|
chr15_+_62359175
|
3.711
|
NM_207322
|
C2CD4A
|
C2 calcium-dependent domain containing 4A
|
|
chr18_+_56807115
|
3.605
|
NM_033280
|
SEC11C
|
SEC11 homolog C (S. cerevisiae)
|
|
chr6_+_37137882
|
3.585
|
NM_001243186
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr19_-_12833461
|
3.584
|
NM_001136196
|
TNPO2
|
transportin 2
|
|
chr15_-_40213087
|
3.501
|
NM_007223
|
GPR176
|
G protein-coupled receptor 176
|
|
chr7_-_22396511
|
3.494
|
NM_012294
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5
|
|
chr1_+_101361688
|
3.434
|
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7
|
|
chr8_-_104427268
|
3.412
|
|
SLC25A32
|
solute carrier family 25, member 32
|
|
chr5_-_121413967
|
3.404
|
|
LOX
|
lysyl oxidase
|
|
chr6_-_82461857
|
3.268
|
|
FAM46A
|
family with sequence similarity 46, member A
|
|
chr14_+_105155724
|
3.264
|
NM_001031714
NM_022489
NM_032714
|
INF2
|
inverted formin, FH2 and WH2 domain containing
|
|
chr5_+_133984234
|
3.258
|
NM_001252231
NM_021982
|
SEC24A
|
SEC24 family, member A (S. cerevisiae)
|
|
chr1_+_154193757
|
3.249
|
|
UBAP2L
|
ubiquitin associated protein 2-like
|
|
chr2_+_208576239
|
3.235
|
NM_001142300
NM_152523
|
CCNYL1
|
cyclin Y-like 1
|
|
chr11_-_85780105
|
3.186
|
NM_001008660
NM_001206946
NM_007166
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr5_-_133706717
|
3.176
|
|
CDKL3
|
cyclin-dependent kinase-like 3
|
|
chr17_-_40264687
|
3.155
|
NM_024119
|
DHX58
|
DEXH (Asp-Glu-X-His) box polypeptide 58
|
|
chr1_-_11120081
|
3.104
|
NM_003132
|
SRM
|
spermidine synthase
|
|
chr5_+_176730812
|
3.077
|
NM_013237
|
PRELID1
|
PRELI domain containing 1
|
|
chr12_-_58146118
|
3.071
|
NM_000075
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr7_+_4815258
|
3.022
|
NM_014855
|
KIAA0415
|
KIAA0415
|
|
chr12_-_58146110
|
3.015
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr1_-_241520467
|
3.014
|
NM_002924
|
RGS7
|
regulator of G-protein signaling 7
|
|
chr5_+_133706869
|
3.011
|
NM_003337
|
UBE2B
|
ubiquitin-conjugating enzyme E2B
|
|
chr5_+_149865310
|
2.994
|
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1
|
|
chr6_+_138188352
|
2.971
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr3_+_133292714
|
2.944
|
|
CDV3
|
CDV3 homolog (mouse)
|
|
chr2_-_220252569
|
2.932
|
|
DNPEP
|
aspartyl aminopeptidase
|
|
chr12_-_58146108
|
2.917
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr12_-_58146025
|
2.911
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr1_-_32281577
|
2.898
|
NM_144569
|
SPOCD1
|
SPOC domain containing 1
|
|
chr20_+_61273786
|
2.892
|
NM_016354
|
SLCO4A1
|
solute carrier organic anion transporter family, member 4A1
|
|
chr16_-_4466607
|
2.867
|
|
CORO7
|
coronin 7
|
|
chr17_-_54991399
|
2.863
|
NM_005082
|
TRIM25
|
tripartite motif containing 25
|
|
chr8_-_8750739
|
2.852
|
NM_004225
|
MFHAS1
|
malignant fibrous histiocytoma amplified sequence 1
|
|
chr19_-_40854283
|
2.841
|
NM_178830
|
C19orf47
|
chromosome 19 open reading frame 47
|
|
chr5_+_70883137
|
2.841
|
|
MCCC2
|
methylcrotonoyl-CoA carboxylase 2 (beta)
|
|
chr5_-_121414011
|
2.829
|
NM_002317
|
LOX
|
lysyl oxidase
|
|
chr10_+_60028885
|
2.820
|
NM_018464
|
CISD1
|
CDGSH iron sulfur domain 1
|
|
chr14_-_65569056
|
2.814
|
|
MAX
|
MYC associated factor X
|
|
chr11_-_85780088
|
2.808
|
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr17_-_63052884
|
2.760
|
NM_006572
|
GNA13
|
guanine nucleotide binding protein (G protein), alpha 13
|
|
chr16_-_1401765
|
2.744
|
NM_001001410
|
C16orf42
|
chromosome 16 open reading frame 42
|
|
chr1_-_154192882
|
2.742
|
|
C1orf43
|
chromosome 1 open reading frame 43
|
|
chr1_-_11120047
|
2.733
|
|
SRM
|
spermidine synthase
|
|
chr1_-_111743261
|
2.729
|
NM_024901
|
DENND2D
|
DENN/MADD domain containing 2D
|
|
chr19_-_12832923
|
2.726
|
NM_013433
|
TNPO2
|
transportin 2
|
|
chr9_+_37079882
|
2.681
|
|
LOC100506710
|
endogenous Bornavirus-like nucleoprotein 2 pseudogene
|
|
chr1_-_212873104
|
2.668
|
|
BATF3
|
basic leucine zipper transcription factor, ATF-like 3
|
|
chr8_+_22224758
|
2.666
|
NM_001128431
NM_001135154
NM_015359
|
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14
|
|
chr3_+_133293256
|
2.653
|
NM_001134423
|
CDV3
|
CDV3 homolog (mouse)
|
|
chr12_+_6833425
|
2.649
|
NM_001164093
|
COPS7A
|
COP9 constitutive photomorphogenic homolog subunit 7A (Arabidopsis)
|
|
chr9_+_74920329
|
2.630
|
|
|
|
|
chr14_-_65569221
|
2.626
|
NM_002382
NM_145112
NM_145113
NM_145114
NM_145116
NM_197957
|
MAX
|
MYC associated factor X
|
|
chr10_-_14050453
|
2.624
|
|
FRMD4A
|
FERM domain containing 4A
|
|
chr3_+_113465901
|
2.613
|
|
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A
|
|
chr20_+_61273823
|
2.578
|
|
SLCO4A1
|
solute carrier organic anion transporter family, member 4A1
|
|
chr1_-_111746917
|
2.573
|
|
DENND2D
|
DENN/MADD domain containing 2D
|
|
chr5_-_121413906
|
2.563
|
|
LOX
|
lysyl oxidase
|
|
chr18_+_77866914
|
2.546
|
NM_014913
|
ADNP2
|
ADNP homeobox 2
|
|
chr1_+_110453456
|
2.510
|
|
CSF1
|
colony stimulating factor 1 (macrophage)
|
|
chr19_+_45682002
|
2.507
|
NM_212550
|
BLOC1S3
|
biogenesis of lysosomal organelles complex-1, subunit 3
|
|
chr6_-_134497069
|
2.471
|
NM_001143678
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr1_-_241520384
|
2.439
|
|
RGS7
|
regulator of G-protein signaling 7
|
|
chr8_+_104427659
|
2.427
|
|
DCAF13
|
DDB1 and CUL4 associated factor 13
|
|
chr9_-_34048887
|
2.424
|
NM_018449
|
UBAP2
|
ubiquitin associated protein 2
|
|
chr1_-_205290756
|
2.415
|
NM_030952
|
NUAK2
|
NUAK family, SNF1-like kinase, 2
|
|
chr15_-_50978921
|
2.415
|
|
TRPM7
|
transient receptor potential cation channel, subfamily M, member 7
|
|
chr12_+_53645906
|
2.401
|
|
MFSD5
|
major facilitator superfamily domain containing 5
|
|
chr12_+_27485767
|
2.395
|
NM_001248002
NM_001248003
NM_001248004
NM_001248005
NM_020183
|
ARNTL2
|
aryl hydrocarbon receptor nuclear translocator-like 2
|
|
chr3_+_119187782
|
2.378
|
NM_152305
|
POGLUT1
|
protein O-glucosyltransferase 1
|
|
chrX_-_101397370
|
2.362
|
NM_001006938
|
TCEAL6
|
transcription elongation factor A (SII)-like 6
|
|
chr8_+_22224786
|
2.361
|
|
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14
|
|
chr2_-_220252513
|
2.332
|
|
DNPEP
|
aspartyl aminopeptidase
|
|
chr5_+_70883100
|
2.325
|
NM_022132
|
MCCC2
|
methylcrotonoyl-CoA carboxylase 2 (beta)
|
|
chr19_-_47290541
|
2.320
|
NM_001145144
|
SLC1A5
|
solute carrier family 1 (neutral amino acid transporter), member 5
|
|
chr17_+_1628032
|
2.293
|
NM_001163809
|
WDR81
|
WD repeat domain 81
|
|
chr12_+_53645869
|
2.281
|
NM_032889
|
MFSD5
|
major facilitator superfamily domain containing 5
|
|
chr18_+_77867145
|
2.253
|
|
ADNP2
|
ADNP homeobox 2
|
|
chr11_-_61684981
|
2.247
|
NM_013401
|
RAB3IL1
|
RAB3A interacting protein (rabin3)-like 1
|
|
chr1_+_166808721
|
2.245
|
NM_017542
|
POGK
|
pogo transposable element with KRAB domain
|
|
chr8_+_123793988
|
2.242
|
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr19_+_7587436
|
2.240
|
NM_020533
|
MCOLN1
|
mucolipin 1
|
|
chr14_+_55168831
|
2.239
|
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr7_+_4815277
|
2.238
|
|
KIAA0415
|
KIAA0415
|
|
chr1_-_53164014
|
2.219
|
|
SELRC1
|
Sel1 repeat containing 1
|
|
chr1_+_183605207
|
2.205
|
NM_015149
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr3_+_184032319
|
2.196
|
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr6_+_151773421
|
2.196
|
NM_024573
|
C6orf211
|
chromosome 6 open reading frame 211
|
|
chr8_+_104427612
|
2.193
|
|
DCAF13
|
DDB1 and CUL4 associated factor 13
|
|
chr15_-_50978960
|
2.186
|
|
TRPM7
|
transient receptor potential cation channel, subfamily M, member 7
|
|
chr11_-_85779821
|
2.180
|
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr12_+_6833264
|
2.172
|
|
COPS7A
|
COP9 constitutive photomorphogenic homolog subunit 7A (Arabidopsis)
|
|
chr3_-_113465101
|
2.172
|
NM_025146
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit
|
|
chr22_-_18256748
|
2.164
|
NM_001244572
NM_197966
|
BID
|
BH3 interacting domain death agonist
|
|
chr17_+_6915735
|
2.160
|
NM_001004333
|
RNASEK
|
ribonuclease, RNase K
|
|
chr8_+_123793894
|
2.151
|
NM_014943
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr1_+_180123975
|
2.147
|
|
QSOX1
|
quiescin Q6 sulfhydryl oxidase 1
|
|
chr7_+_916178
|
2.136
|
NM_015949
|
GET4
|
golgi to ER traffic protein 4 homolog (S. cerevisiae)
|
|
chr17_+_7211693
|
2.134
|
NM_001143762
|
EIF5A
|
eukaryotic translation initiation factor 5A
|
|
chr8_+_22224997
|
2.127
|
NM_001135153
|
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14
|
|
chr16_-_4466595
|
2.114
|
|
CORO7
|
coronin 7
|
|
chr2_-_193059599
|
2.113
|
NM_016192
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chr2_-_10588273
|
2.105
|
|
ODC1
|
ornithine decarboxylase 1
|
|
chr12_+_6833149
|
2.102
|
NM_001164094
NM_016319
NM_001164095
|
COPS7A
|
COP9 constitutive photomorphogenic homolog subunit 7A (Arabidopsis)
|
|
chr15_-_50978983
|
2.085
|
NM_017672
|
TRPM7
|
transient receptor potential cation channel, subfamily M, member 7
|
|
chr12_+_6833288
|
2.073
|
|
COPS7A
|
COP9 constitutive photomorphogenic homolog subunit 7A (Arabidopsis)
|
|
chr12_-_57472487
|
2.069
|
|
TMEM194A
|
transmembrane protein 194A
|
|
chr1_+_101361589
|
2.051
|
NM_001144884
NM_133496
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7
|
|
chr18_+_72163582
|
2.047
|
|
CNDP2
|
CNDP dipeptidase 2 (metallopeptidase M20 family)
|
|
chr8_-_104427319
|
2.041
|
|
SLC25A32
|
solute carrier family 25, member 32
|
|
chr5_+_34656595
|
2.039
|
NM_001145520
|
RAI14
|
retinoic acid induced 14
|
|
chr11_-_59436488
|
2.038
|
NM_152716
|
PATL1
|
protein associated with topoisomerase II homolog 1 (yeast)
|
|
chr1_-_159895279
|
2.035
|
NM_003564
|
TAGLN2
|
transgelin 2
|
|
chr17_-_63052744
|
2.032
|
|
GNA13
|
guanine nucleotide binding protein (G protein), alpha 13
|
|
chr4_-_15656974
|
2.031
|
NM_001193534
NM_001193535
NM_012161
|
FBXL5
|
F-box and leucine-rich repeat protein 5
|
|
chr1_+_101361671
|
2.028
|
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7
|
|
chr1_-_53163985
|
2.027
|
|
SELRC1
|
Sel1 repeat containing 1
|
|
chr12_-_49318663
|
2.026
|
NM_001143781
|
FKBP11
|
FK506 binding protein 11, 19 kDa
|
|
chr16_+_69984625
|
2.026
|
NM_001136214
|
CLEC18A
|
C-type lectin domain family 18, member A
|
|
chr12_+_132413776
|
2.025
|
NM_001002019
NM_025215
|
PUS1
|
pseudouridylate synthase 1
|
|
chr6_-_28891409
|
2.012
|
|
TRIM27
|
tripartite motif containing 27
|
|
chr1_-_31712733
|
2.011
|
NM_024522
|
NKAIN1
|
Na+/K+ transporting ATPase interacting 1
|
|
chr7_-_2354010
|
2.001
|
|
SNX8
|
sorting nexin 8
|
|
chr12_+_53645985
|
2.001
|
|
MFSD5
|
major facilitator superfamily domain containing 5
|
|
chr1_-_52831817
|
1.991
|
NM_032449
|
CC2D1B
|
coiled-coil and C2 domain containing 1B
|
|
chr1_+_180124013
|
1.984
|
|
QSOX1
|
quiescin Q6 sulfhydryl oxidase 1
|
|
chr15_-_50978726
|
1.984
|
|
TRPM7
|
transient receptor potential cation channel, subfamily M, member 7
|
|
chr12_-_57472547
|
1.972
|
|
TMEM194A
|
transmembrane protein 194A
|
|
chr17_-_18218203
|
1.969
|
NM_004618
|
TOP3A
|
topoisomerase (DNA) III alpha
|
|
chr11_-_126081532
|
1.969
|
NM_001144827
NM_032795
|
RPUSD4
|
RNA pseudouridylate synthase domain containing 4
|
|
chr8_+_22102845
|
1.962
|
|
POLR3D
|
polymerase (RNA) III (DNA directed) polypeptide D, 44kDa
|
|
chr19_-_48018163
|
1.959
|
|
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha
|
|
chr1_-_111743297
|
1.959
|
|
DENND2D
|
DENN/MADD domain containing 2D
|
|
chr16_+_70613797
|
1.957
|
NM_001172771
NM_152456
|
IL34
|
interleukin 34
|
|
chr16_-_4466570
|
1.954
|
|
CORO7-PAM16
CORO7
|
CORO7-PAM16 readthrough
coronin 7
|
|
chr10_+_6186903
|
1.951
|
|
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
|
|
chrX_+_128913897
|
1.950
|
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chr5_+_176730779
|
1.950
|
|
PRELID1
|
PRELI domain containing 1
|
|
chr7_-_47621635
|
1.947
|
|
TNS3
|
tensin 3
|
|
chr16_+_56965747
|
1.941
|
NM_001010989
NM_001010990
NM_014685
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1
|
|
chr17_-_41623652
|
1.941
|
NM_001079675
|
ETV4
|
ets variant 4
|
|
chr17_+_80332152
|
1.931
|
NM_018949
|
UTS2R
|
urotensin 2 receptor
|
|
chr19_+_10812111
|
1.931
|
NM_031209
|
QTRT1
|
queuine tRNA-ribosyltransferase 1
|