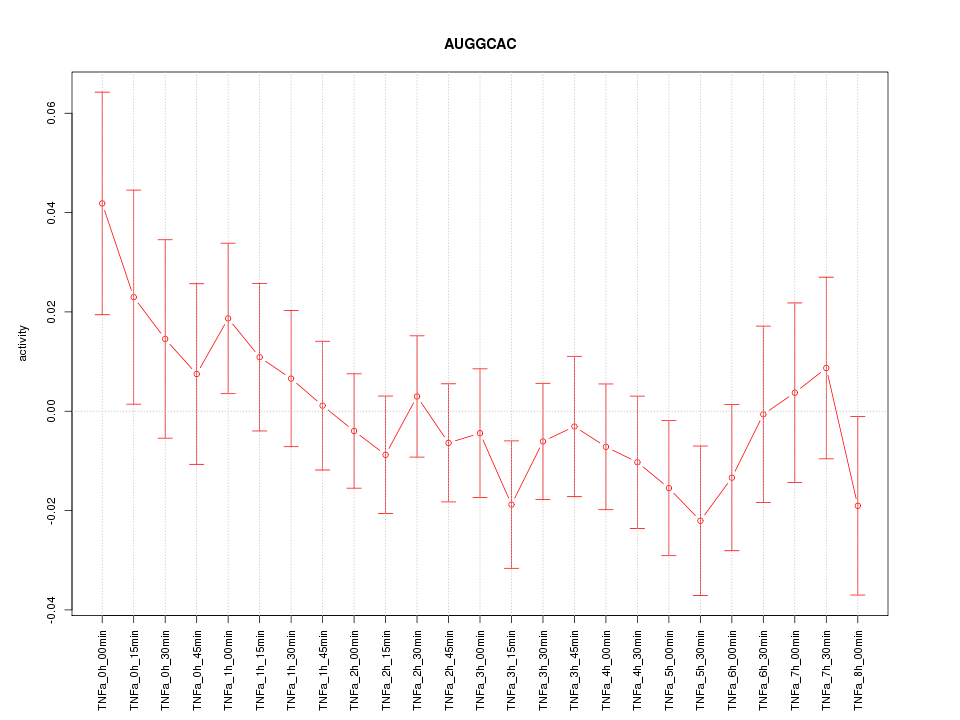
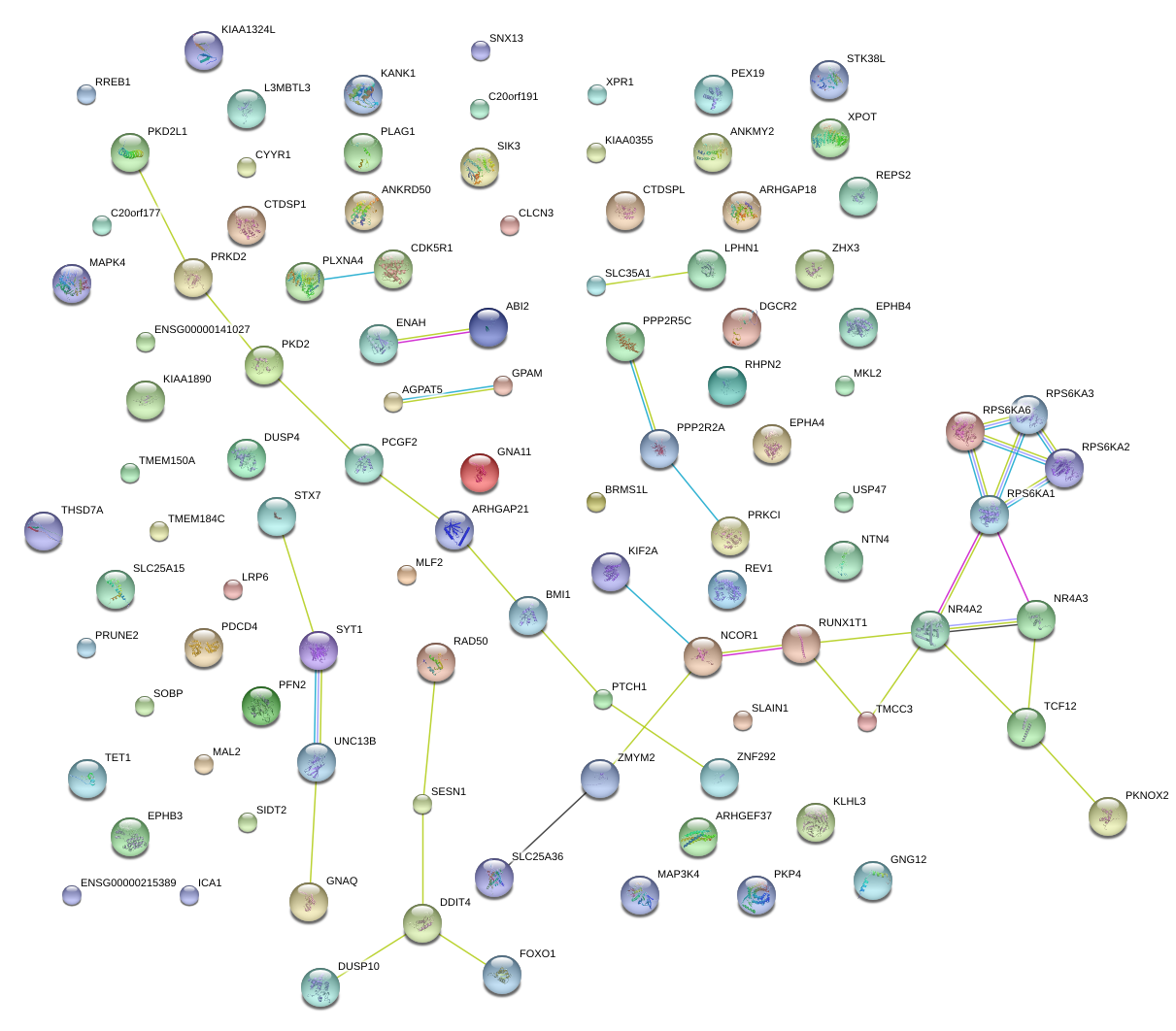
|
chr12_-_95044205
|
3.046
|
NM_020698
|
TMCC3
|
transmembrane and coiled-coil domain family 3
|
|
chr8_+_120220554
|
2.997
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr6_-_130031278
|
2.427
|
NM_033515
|
ARHGAP18
|
Rho GTPase activating protein 18
|
|
chr3_+_37903643
|
2.316
|
NM_001008392
NM_005808
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like
|
|
chr10_+_112631552
|
2.303
|
NM_001199492
NM_014456
NM_145341
|
PDCD4
|
programmed cell death 4 (neoplastic transformation inhibitor)
|
|
chr12_+_27397077
|
2.226
|
NM_015000
|
STK38L
|
serine/threonine kinase 38 like
|
|
chr5_+_148961035
|
1.885
|
NM_001001669
|
ARHGEF37
|
Rho guanine nucleotide exchange factor (GEF) 37
|
|
chr8_+_6565870
|
1.696
|
NM_018361
|
AGPAT5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon)
|
|
chr15_+_57210823
|
1.564
|
NM_003205
NM_207036
NM_207037
NM_207038
|
TCF12
|
transcription factor 12
|
|
chr20_+_58508818
|
1.488
|
NM_001190826
|
C20orf177
|
chromosome 20 open reading frame 177
|
|
chr8_-_4852262
|
1.471
|
NM_033225
|
CSMD1
|
CUB and Sushi multiple domains 1
|
|
chr10_+_70320044
|
1.452
|
NM_030625
|
TET1
|
tet methylcytosine dioxygenase 1
|
|
chr20_+_58515408
|
1.445
|
NM_001190827
NM_022106
|
C20orf177
|
chromosome 20 open reading frame 177
|
|
chr6_+_161412807
|
1.428
|
NM_005922
NM_006724
|
MAP3K4
|
mitogen-activated protein kinase kinase kinase 4
|
|
chr13_+_78271820
|
1.410
|
NM_001242868
|
SLAIN1
|
SLAIN motif family, member 1
|
|
chr4_+_88928797
|
1.386
|
NM_000297
|
PKD2
|
polycystic kidney disease 2 (autosomal dominant)
|
|
chrX_-_20284708
|
1.364
|
NM_004586
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3
|
|
chr15_+_57511616
|
1.290
|
NM_207040
|
TCF12
|
transcription factor 12
|
|
chr9_-_79520878
|
1.247
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr13_+_78273015
|
1.231
|
NM_001040153
|
SLAIN1
|
SLAIN motif family, member 1
|
|
chr8_-_93029864
|
1.219
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr21_-_27945267
|
1.193
|
NM_052954
|
CYYR1
|
cysteine/tyrosine-rich 1
|
|
chr8_-_57123858
|
1.153
|
NM_001114634
NM_001114635
NM_002655
|
PLAG1
|
pleiomorphic adenoma gene 1
|
|
chr17_+_30813928
|
1.152
|
NM_003885
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35)
|
|
chr8_-_93107881
|
1.137
|
NM_001198625
NM_001198626
NM_001198627
NM_001198631
NM_001198634
NM_001198679
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_-_93111915
|
1.129
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr7_-_16685360
|
1.116
|
NM_020319
|
ANKMY2
|
ankyrin repeat and MYND domain containing 2
|
|
chr8_-_93107442
|
1.100
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr7_-_11871735
|
1.078
|
NM_015204
|
THSD7A
|
thrombospondin, type I, domain containing 7A
|
|
chr12_-_12419702
|
1.067
|
NM_002336
|
LRP6
|
low density lipoprotein receptor-related protein 6
|
|
chr8_-_93115453
|
1.067
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr13_+_78315294
|
1.064
|
NM_001242869
NM_001242870
NM_001242871
NM_144595
|
SLAIN1
|
SLAIN motif family, member 1
|
|
chr6_-_109330190
|
1.051
|
NM_001199934
|
SESN1
|
sestrin 1
|
|
chr2_+_159313466
|
1.010
|
NM_001005476
NM_003628
|
PKP4
|
plakophilin 4
|
|
chr6_+_88182624
|
0.990
|
NM_001168398
NM_006416
|
SLC35A1
|
solute carrier family 35 (CMP-sialic acid transporter), member A1
|
|
chr8_-_93075190
|
0.943
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr2_+_219263060
|
0.931
|
NM_001206878
|
CTDSP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1
|
|
chr6_-_109415514
|
0.908
|
NM_014454
|
SESN1
|
sestrin 1
|
|
chr6_-_109330752
|
0.902
|
NM_001199933
|
SESN1
|
sestrin 1
|
|
chr2_+_219264477
|
0.885
|
NM_021198
NM_182642
|
CTDSP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1
|
|
chr7_-_8301767
|
0.852
|
NM_001136020
|
ICA1
|
islet cell autoantigen 1, 69kDa
|
|
chr6_+_7107986
|
0.848
|
NM_001003698
NM_001003699
NM_001003700
|
RREB1
|
ras responsive element binding protein 1
|
|
chr4_-_125633716
|
0.845
|
NM_001167882
NM_020337
|
ANKRD50
|
ankyrin repeat domain 50
|
|
chr2_+_204192915
|
0.842
|
NM_005759
|
ABI2
|
abl-interactor 2
|
|
chr12_-_96184510
|
0.836
|
NM_021229
|
NTN4
|
netrin 4
|
|
chr13_-_41240607
|
0.829
|
NM_002015
|
FOXO1
|
forkhead box O1
|
|
chr2_-_157189040
|
0.828
|
NM_006186
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chr9_+_706744
|
0.820
|
NM_153186
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr9_+_504661
|
0.812
|
NM_015158
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr10_-_113943467
|
0.790
|
NM_001244949
NM_020918
|
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial
|
|
chr11_+_125034558
|
0.775
|
NM_022062
|
PKNOX2
|
PBX/knotted 1 homeobox 2
|
|
chr6_+_7107799
|
0.764
|
NM_001168344
|
RREB1
|
ras responsive element binding protein 1
|
|
chr3_-_149688638
|
0.760
|
NM_002628
NM_053024
|
PFN2
|
profilin 2
|
|
chr19_+_34745452
|
0.747
|
NM_014686
|
KIAA0355
|
KIAA0355
|
|
chr12_+_79258448
|
0.737
|
NM_005639
|
SYT1
|
synaptotagmin I
|
|
chr2_-_222436996
|
0.730
|
NM_004438
|
EPHA4
|
EPH receptor A4
|
|
chr22_-_19109912
|
0.727
|
NM_001173533
NM_001173534
NM_001184781
NM_005137
|
DGCR2
|
DiGeorge syndrome critical region gene 2
|
|
chr12_+_79439432
|
0.722
|
NM_001135806
|
SYT1
|
synaptotagmin I
|
|
chr12_+_79257772
|
0.722
|
NM_001135805
|
SYT1
|
synaptotagmin I
|
|
chr10_+_74033676
|
0.709
|
NM_019058
|
DDIT4
|
DNA-damage-inducible transcript 4
|
|
chr13_+_41363546
|
0.704
|
NM_014252
|
SLC25A15
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15
|
|
chr14_+_102228134
|
0.704
|
NM_001161725
NM_001161726
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr19_-_14316980
|
0.686
|
NM_001008701
NM_014921
|
LPHN1
|
latrophilin 1
|
|
chr4_+_148538508
|
0.685
|
NM_018241
|
TMEM184C
|
transmembrane protein 184C
|
|
chr7_-_8301681
|
0.682
|
NM_022307
|
ICA1
|
islet cell autoantigen 1, 69kDa
|
|
chr6_-_132834165
|
0.680
|
NM_003569
|
STX7
|
syntaxin 7
|
|
chr8_+_26149004
|
0.668
|
NM_002717
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chr6_+_107811272
|
0.667
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr5_+_131892556
|
0.651
|
NM_005732
|
RAD50
|
RAD50 homolog (S. cerevisiae)
|
|
chr2_-_85829757
|
0.640
|
NM_001031738
|
TMEM150A
|
transmembrane protein 150A
|
|
chr13_+_20532788
|
0.626
|
NM_003453
NM_197968
NM_001190964
|
ZMYM2
|
zinc finger, MYM-type 2
|
|
chr11_-_116968986
|
0.611
|
NM_025164
|
SIK3
|
SIK family kinase 3
|
|
chr7_-_132261268
|
0.609
|
NM_001105543
NM_020911
|
PLXNA4
|
plexin A4
|
|
chr8_-_29206321
|
0.607
|
NM_057158
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr4_+_170541656
|
0.606
|
NM_001243372
NM_001829
NM_173872
|
CLCN3
|
chloride channel 3
|
|
chr10_-_25012543
|
0.605
|
NM_020824
|
ARHGAP21
|
Rho GTPase activating protein 21
|
|
chr8_+_26150731
|
0.600
|
NM_001177591
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chr9_-_80646150
|
0.596
|
NM_002072
|
GNAQ
|
guanine nucleotide binding protein (G protein), q polypeptide
|
|
chr11_+_117049838
|
0.589
|
NM_001040455
|
SIDT2
|
SID1 transmembrane family, member 2
|
|
chr1_-_221915458
|
0.581
|
NM_007207
NM_144729
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr13_+_20532906
|
0.569
|
NM_001190965
|
ZMYM2
|
zinc finger, MYM-type 2
|
|
chr1_-_225840660
|
0.568
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr3_+_169940214
|
0.564
|
NM_002740
|
PRKCI
|
protein kinase C, iota
|
|
chr8_-_29207795
|
0.557
|
NM_001394
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr5_-_137071703
|
0.557
|
NM_017415
|
KLHL3
|
kelch-like 3 (Drosophila)
|
|
chr2_-_100106478
|
0.556
|
NM_001037872
NM_016316
|
REV1
|
REV1 homolog (S. cerevisiae)
|
|
chr19_-_33555757
|
0.555
|
NM_033103
|
RHPN2
|
rhophilin, Rho GTPase binding protein 2
|
|
chr9_+_35161950
|
0.539
|
NM_006377
|
UNC13B
|
unc-13 homolog B (C. elegans)
|
|
chr1_-_68299047
|
0.530
|
NM_018841
|
GNG12
|
guanine nucleotide binding protein (G protein), gamma 12
|
|
chr10_+_22610138
|
0.522
|
NM_005180
|
BMI1
|
BMI1 polycomb ring finger oncogene
|
|
chr1_+_180601121
|
0.516
|
NM_001135669
NM_004736
|
XPR1
|
xenotropic and polytropic retrovirus receptor 1
|
|
chr5_+_61601944
|
0.500
|
NM_001098511
NM_001243952
NM_001243953
NM_004520
|
KIF2A
|
kinesin heavy chain member 2A
|
|
chr12_+_64798050
|
0.497
|
NM_007235
|
XPOT
|
exportin, tRNA (nuclear export receptor for tRNAs)
|
|
chr1_-_221910801
|
0.495
|
NM_144728
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr7_-_8302141
|
0.493
|
NM_004968
|
ICA1
|
islet cell autoantigen 1, 69kDa
|
|
chr9_-_98269480
|
0.486
|
NM_001083607
|
PTCH1
|
patched 1
|
|
chr3_+_140660639
|
0.480
|
NM_001104647
NM_018155
|
SLC25A36
|
solute carrier family 25, member 36
|
|
chr7_-_17980102
|
0.476
|
NM_015132
|
SNX13
|
sorting nexin 13
|
|
chr14_+_36295515
|
0.472
|
NM_032352
|
BRMS1L
|
breast cancer metastasis-suppressor 1-like
|
|
chr9_-_98270321
|
0.460
|
NM_001083604
NM_001083605
|
PTCH1
|
patched 1
|
|
chr6_+_87865261
|
0.456
|
NM_015021
|
ZNF292
|
zinc finger protein 292
|
|
chr17_-_16118789
|
0.455
|
NM_006311
|
NCOR1
|
nuclear receptor corepressor 1
|
|
chr16_+_14164879
|
0.448
|
NM_014048
|
MKL2
|
MKL/myocardin-like 2
|
|
chr9_-_98270830
|
0.439
|
NM_000264
|
PTCH1
|
patched 1
|
|
chr14_+_102276125
|
0.431
|
NM_002719
NM_178586
NM_178587
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chrX_+_16964730
|
0.407
|
NM_001080975
NM_004726
|
REPS2
|
RALBP1 associated Eps domain containing 2
|
|
chr1_-_160254923
|
0.404
|
NM_001193644
NM_002857
|
PEX19
|
peroxisomal biogenesis factor 19
|
|
chr7_-_86595203
|
0.381
|
NM_152748
|
KIAA1324L
|
KIAA1324-like
|
|
chr20_-_39928708
|
0.377
|
NM_015035
|
ZHX3
|
zinc fingers and homeoboxes 3
|
|
chr6_+_130339715
|
0.377
|
NM_001007102
NM_032438
|
L3MBTL3
|
l(3)mbt-like 3 (Drosophila)
|
|
chr11_+_11862955
|
0.365
|
NM_017944
|
USP47
|
ubiquitin specific peptidase 47
|
|
chr18_+_48086402
|
0.364
|
NM_002747
|
MAPK4
|
mitogen-activated protein kinase 4
|
|
chr4_-_78740508
|
0.363
|
NM_144571
|
CNOT6L
|
CCR4-NOT transcription complex, subunit 6-like
|
|
chr4_-_129209983
|
0.358
|
NM_006320
|
PGRMC2
|
progesterone receptor membrane component 2
|
|
chrY_+_15815446
|
0.346
|
NM_004202
|
TMSB4Y
|
thymosin beta 4, Y-linked
|
|
chr12_+_98909331
|
0.341
|
NM_001032283
NM_001032284
NM_003276
|
TMPO
|
thymopoietin
|
|
chr1_+_185014449
|
0.334
|
NM_007212
|
RNF2
|
ring finger protein 2
|
|
chrX_+_12993224
|
0.326
|
NM_021109
|
TMSB4X
|
thymosin beta 4, X-linked
|
|
chr19_+_34287662
|
0.320
|
NM_001129994
NM_001129995
NM_024076
|
KCTD15
|
potassium channel tetramerisation domain containing 15
|
|
chr7_-_86688944
|
0.319
|
NM_001142749
|
KIAA1324L
|
KIAA1324-like
|
|
chr9_-_107690526
|
0.318
|
NM_005502
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr5_+_271700
|
0.312
|
NM_013232
|
PDCD6
|
programmed cell death 6
|
|
chr1_+_203595897
|
0.294
|
NM_001001396
NM_001684
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4
|
|
chr15_+_64388082
|
0.292
|
NM_001242933
NM_003099
NM_148955
|
SNX1
|
sorting nexin 1
|
|
chr9_-_98269686
|
0.286
|
NM_001083606
|
PTCH1
|
patched 1
|
|
chr22_-_21356351
|
0.285
|
NM_001008695
NM_030573
|
THAP7
|
THAP domain containing 7
|
|
chr18_-_53255734
|
0.276
|
NM_001083962
NM_001243230
NM_003199
|
TCF4
|
transcription factor 4
|
|
chr6_-_7910278
|
0.267
|
NM_001145549
|
TXNDC5
|
thioredoxin domain containing 5 (endoplasmic reticulum)
|
|
chr14_-_35183722
|
0.266
|
NM_001243645
NM_138638
|
CFL2
|
cofilin 2 (muscle)
|
|
chr22_+_40440664
|
0.265
|
NM_001024843
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr2_+_206547131
|
0.261
|
NM_003872
NM_018534
NM_201264
NM_201266
NM_201267
NM_201279
|
NRP2
|
neuropilin 2
|
|
chr14_+_53196852
|
0.257
|
NM_001130701
NM_145251
|
STYX
|
serine/threonine/tyrosine interacting protein
|
|
chr7_-_129592794
|
0.257
|
NM_003344
NM_182697
|
UBE2H
|
ubiquitin-conjugating enzyme E2H
|
|
chr5_+_102201526
|
0.250
|
NM_000919
NM_001177306
NM_138766
NM_138821
NM_138822
|
PAM
|
peptidylglycine alpha-amidating monooxygenase
|
|
chr5_-_77944341
|
0.250
|
NM_005779
|
LHFPL2
|
lipoma HMGIC fusion partner-like 2
|
|
chr1_+_70671331
|
0.244
|
NM_001190987
NM_004768
|
SRSF11
|
serine/arginine-rich splicing factor 11
|
|
chr3_-_125094003
|
0.243
|
NM_021964
|
ZNF148
|
zinc finger protein 148
|
|
chr4_-_114682836
|
0.237
|
NM_001221
NM_172114
NM_172115
NM_172127
NM_172128
NM_172129
|
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta
|
|
chr12_-_92539614
|
0.236
|
NM_001731
|
BTG1
|
B-cell translocation gene 1, anti-proliferative
|
|
chr8_+_97274158
|
0.235
|
NM_014754
|
PTDSS1
|
phosphatidylserine synthase 1
|
|
chr17_-_36956145
|
0.233
|
NM_003559
|
PIP4K2B
|
phosphatidylinositol-5-phosphate 4-kinase, type II, beta
|
|
chr22_+_40573879
|
0.229
|
NM_001162501
NM_015088
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr14_-_88737160
|
0.222
|
NM_138318
|
KCNK10
|
potassium channel, subfamily K, member 10
|
|
chr7_+_35840595
|
0.222
|
NM_001011553
NM_001242956
NM_001788
|
SEPT7
|
septin 7
|
|
chr7_-_129592711
|
0.213
|
NM_001202498
|
UBE2H
|
ubiquitin-conjugating enzyme E2H
|
|
chr14_-_35182944
|
0.196
|
NM_021914
|
CFL2
|
cofilin 2 (muscle)
|
|
chr5_+_72251753
|
0.191
|
NM_001146032
NM_138782
|
FCHO2
|
FCH domain only 2
|
|
chr14_+_105886146
|
0.190
|
NM_001203258
NM_004689
|
MTA1
|
metastasis associated 1
|
|
chr5_-_36242243
|
0.188
|
NM_153013
|
NADKD1
|
NAD kinase domain containing 1
|
|
chr5_+_133861685
|
0.188
|
NM_015288
|
PHF15
|
PHD finger protein 15
|
|
chr17_-_16097882
|
0.184
|
NM_001190438
NM_001190440
|
NCOR1
|
nuclear receptor corepressor 1
|
|
chr2_+_139259349
|
0.180
|
NM_001001664
|
SPOPL
|
speckle-type POZ protein-like
|
|
chr8_+_64081108
|
0.176
|
NM_152758
|
YTHDF3
|
YTH domain family, member 3
|
|
chr14_+_78870092
|
0.174
|
NM_004796
|
NRXN3
|
neurexin 3
|
|
chr10_+_92980322
|
0.174
|
NM_032373
|
PCGF5
|
polycomb group ring finger 5
|
|
chr11_+_125439294
|
0.171
|
NM_001007277
NM_004879
|
EI24
|
etoposide induced 2.4 mRNA
|
|
chr10_+_22605300
|
0.169
|
NM_001204062
NM_012071
|
COMMD3-BMI1
COMMD3
|
COMMD3-BMI1 readthrough
COMM domain containing 3
|
|
chr18_+_29672568
|
0.168
|
NM_001191324
NM_198128
|
RNF138
|
ring finger protein 138
|
|
chr13_+_108870713
|
0.167
|
NM_032859
|
ABHD13
|
abhydrolase domain containing 13
|
|
chr18_+_29671817
|
0.157
|
NM_016271
|
RNF138
|
ring finger protein 138
|
|
chr8_+_26240522
|
0.156
|
NM_004331
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like
|
|
chr6_+_151646634
|
0.156
|
NM_144497
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr5_-_36241899
|
0.155
|
NM_001085411
|
NADKD1
|
NAD kinase domain containing 1
|
|
chr6_+_151561094
|
0.154
|
NM_005100
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr11_+_63655986
|
0.152
|
NM_017490
|
MARK2
|
MAP/microtubule affinity-regulating kinase 2
|
|
chr20_+_1874779
|
0.152
|
NM_001040022
|
SIRPA
|
signal-regulatory protein alpha
|
|
chr19_+_48958765
|
0.151
|
NM_013348
|
KCNJ14
|
potassium inwardly-rectifying channel, subfamily J, member 14
|
|
chr9_-_98279246
|
0.140
|
NM_001083602
NM_001083603
|
PTCH1
|
patched 1
|
|
chr2_-_69870976
|
0.138
|
NM_014911
|
AAK1
|
AP2 associated kinase 1
|
|
chr6_-_7910716
|
0.138
|
NM_030810
|
TXNDC5
|
thioredoxin domain containing 5 (endoplasmic reticulum)
|
|
chr11_+_12695851
|
0.137
|
NM_021961
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor)
|
|
chr6_+_64356430
|
0.129
|
NM_015153
|
PHF3
|
PHD finger protein 3
|
|
chr15_-_65809603
|
0.124
|
NM_197960
|
DPP8
|
dipeptidyl-peptidase 8
|
|
chr1_-_67896098
|
0.108
|
NM_001018067
NM_001018068
NM_001018069
NM_015640
|
SERBP1
|
SERPINE1 mRNA binding protein 1
|
|
chr20_+_15177503
|
0.107
|
NM_001033087
|
MACROD2
|
MACRO domain containing 2
|
|
chr7_-_111846384
|
0.107
|
NM_014705
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr1_-_153895450
|
0.106
|
NM_020699
|
GATAD2B
|
GATA zinc finger domain containing 2B
|
|
chrX_-_20135015
|
0.106
|
NM_001168465
NM_001168466
NM_152780
|
MAP7D2
|
MAP7 domain containing 2
|
|
chr16_+_85645014
|
0.105
|
NM_001134473
|
KIAA0182
|
KIAA0182
|
|
chr3_+_196439211
|
0.105
|
NM_001166304
NM_017861
|
PIGX
|
phosphatidylinositol glycan anchor biosynthesis, class X
|
|
chrX_-_20134749
|
0.104
|
NM_001168467
|
MAP7D2
|
MAP7 domain containing 2
|
|
chr16_+_23568833
|
0.100
|
NM_019116
|
UBFD1
|
ubiquitin family domain containing 1
|
|
chr7_-_71802207
|
0.095
|
NM_001017440
|
CALN1
|
calneuron 1
|
|
chr12_+_93771607
|
0.089
|
NM_019094
NM_199040
|
NUDT4
NUDT4P1
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4
nudix (nucleoside diphosphate linked moiety X)-type motif 4 pseudogene 1
|
|
chr20_+_13976014
|
0.088
|
NM_080676
|
MACROD2
|
MACRO domain containing 2
|
|
chr14_-_91282760
|
0.088
|
NM_001010854
|
TTC7B
|
tetratricopeptide repeat domain 7B
|
|
chr2_+_169312793
|
0.087
|
NM_203463
|
CERS6
|
ceramide synthase 6
|
|
chr15_-_65810020
|
0.084
|
NM_017743
NM_130434
NM_197961
|
DPP8
|
dipeptidyl-peptidase 8
|
|
chr1_-_93257950
|
0.083
|
NM_005665
|
EVI5
|
ecotropic viral integration site 5
|
|
chr12_-_76953561
|
0.078
|
NM_001003712
NM_020841
|
OSBPL8
|
oxysterol binding protein-like 8
|
|
chr11_+_8102691
|
0.076
|
NM_177972
|
TUB
|
tubby homolog (mouse)
|
|
chr15_-_60919642
|
0.075
|
NM_002943
NM_134260
|
RORA
|
RAR-related orphan receptor A
|
|
chr19_+_5623045
|
0.071
|
NM_001201338
NM_001201339
NM_001201340
NM_002967
|
SAFB
|
scaffold attachment factor B
|
|
chr6_+_131148544
|
0.071
|
NM_001195597
|
LOC100507203
|
uncharacterized LOC100507203
|
|
chr3_+_171758289
|
0.065
|
NM_001135095
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr11_+_8060179
|
0.060
|
NM_003320
|
TUB
|
tubby homolog (mouse)
|
|
chr4_+_15004297
|
0.057
|
NM_001177381
NM_001177382
NM_001177383
NM_001177384
NM_182485
NM_182646
|
CPEB2
|
cytoplasmic polyadenylation element binding protein 2
|
|
chr1_-_70820363
|
0.057
|
NM_030816
|
ANKRD13C
|
ankyrin repeat domain 13C
|
|
chr10_+_114709963
|
0.053
|
NM_001146274
NM_001146283
NM_001146284
NM_001146285
NM_001146286
NM_001198525
NM_001198526
NM_001198527
NM_001198528
NM_001198529
NM_001198530
NM_001198531
NM_030756
|
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box)
|
|
chr15_+_85291802
|
0.052
|
NM_014630
|
ZNF592
|
zinc finger protein 592
|