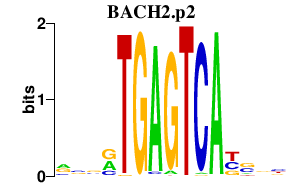
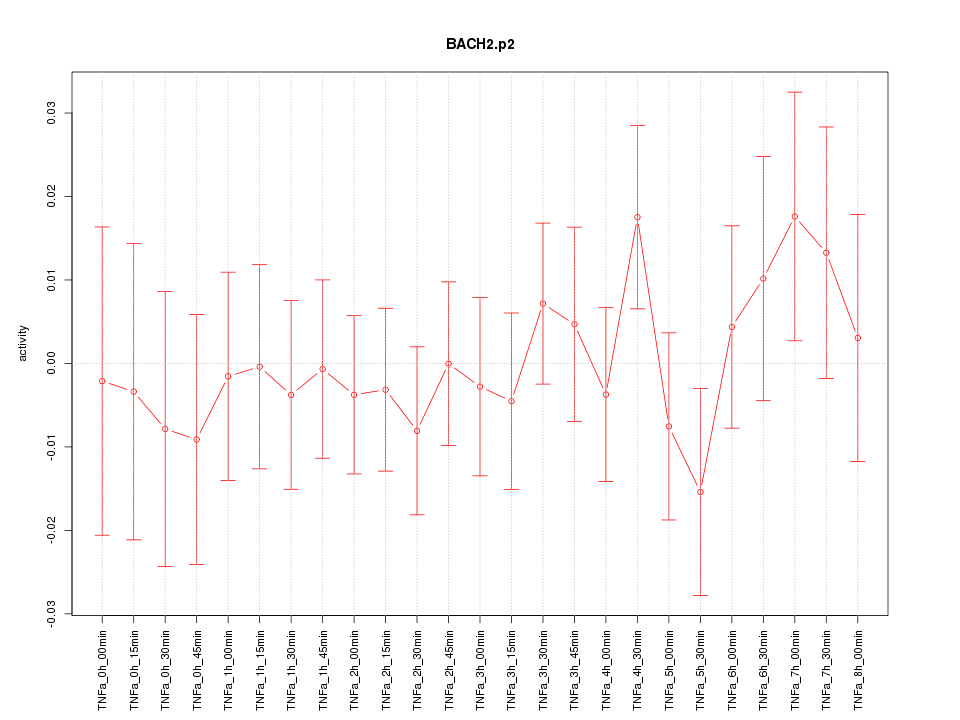
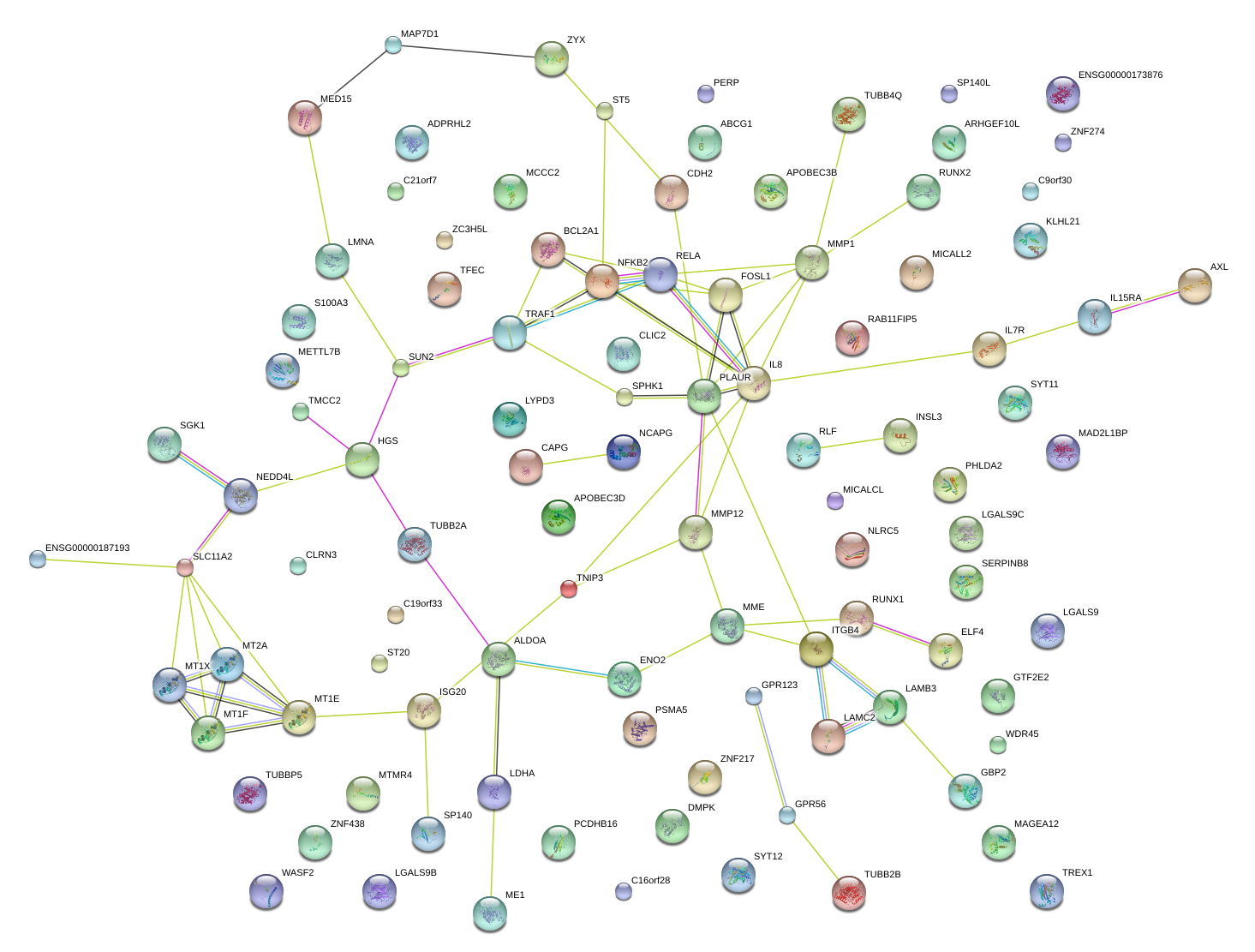
|
chr11_-_102668892
|
3.283
|
NM_001145938
NM_002421
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase)
|
|
chr11_-_102668822
|
3.238
|
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase)
|
|
chr12_+_56075329
|
2.401
|
NM_152637
|
METTL7B
|
methyltransferase like 7B
|
|
chr15_+_89182038
|
2.347
|
NM_002201
|
ISG20
|
interferon stimulated exonuclease gene 20kDa
|
|
chr9_-_123688352
|
2.235
|
|
TRAF1
|
TNF receptor-associated factor 1
|
|
chr1_-_153521717
|
2.037
|
NM_002960
|
S100A3
|
S100 calcium binding protein A3
|
|
chr5_+_35856990
|
1.809
|
NM_002185
|
IL7R
|
interleukin 7 receptor
|
|
chr12_+_56075418
|
1.763
|
|
METTL7B
|
methyltransferase like 7B
|
|
chr1_-_209824641
|
1.743
|
NM_001017402
|
LAMB3
|
laminin, beta 3
|
|
chr15_-_80263460
|
1.558
|
NM_001114735
NM_004049
|
BCL2A1
|
BCL2-related protein A1
|
|
chrX_-_154563735
|
1.552
|
NM_001289
|
CLIC2
|
chloride intracellular channel 2
|
|
chr1_+_183155393
|
1.514
|
|
LAMC2
|
laminin, gamma 2
|
|
chr16_+_57662301
|
1.454
|
NM_001145771
NM_001145772
NM_001145773
NM_201524
NM_201525
|
GPR56
|
G protein-coupled receptor 56
|
|
chr1_+_183155361
|
1.259
|
|
LAMC2
|
laminin, gamma 2
|
|
chr11_-_8739398
|
1.175
|
|
ST5
|
suppression of tumorigenicity 5
|
|
chr10_-_6019454
|
1.163
|
NM_002189
NM_172200
|
IL15RA
|
interleukin 15 receptor, alpha
|
|
chr1_+_156084458
|
1.124
|
NM_005572
NM_170707
NM_170708
|
LMNA
|
lamin A/C
|
|
chr11_+_12308446
|
1.092
|
NM_032867
|
MICALCL
|
MICAL C-terminal like
|
|
chr4_-_122085475
|
0.991
|
NM_024873
|
TNIP3
|
TNFAIP3 interacting protein 3
|
|
chr10_-_31288420
|
0.958
|
NM_001143769
|
ZNF438
|
zinc finger protein 438
|
|
chr6_-_134496833
|
0.954
|
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr7_-_115670774
|
0.932
|
|
TFEC
|
transcription factor EC
|
|
chr17_+_79650961
|
0.931
|
NM_004712
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate
|
|
chr4_+_74606222
|
0.898
|
NM_000584
|
IL8
|
interleukin 8
|
|
chr1_+_183155173
|
0.895
|
NM_005562
NM_018891
|
LAMC2
|
laminin, gamma 2
|
|
chr17_+_79651072
|
0.895
|
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate
|
|
chr19_+_38794800
|
0.859
|
NM_033520
|
C19orf33
|
chromosome 19 open reading frame 33
|
|
chr12_+_7023490
|
0.854
|
NM_001975
|
ENO2
|
enolase 2 (gamma, neuronal)
|
|
chr17_+_79651035
|
0.852
|
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate
|
|
chr19_-_44174497
|
0.841
|
NM_001005376
NM_001005377
NM_002659
|
PLAUR
|
plasminogen activator, urokinase receptor
|
|
chr21_-_36421630
|
0.839
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr10_-_6019880
|
0.809
|
NM_001243539
|
IL15RA
|
interleukin 15 receptor, alpha
|
|
chr11_-_65667809
|
0.808
|
|
FOSL1
|
FOS-like antigen 1
|
|
chr17_+_79651042
|
0.806
|
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate
|
|
chr11_-_2950593
|
0.743
|
NM_003311
|
PHLDA2
|
pleckstrin homology-like domain, family A, member 2
|
|
chr11_-_65667858
|
0.743
|
|
FOSL1
|
FOS-like antigen 1
|
|
chr2_-_85641128
|
0.740
|
|
CAPG
|
capping protein (actin filament), gelsolin-like
|
|
chr1_-_6662681
|
0.725
|
|
KLHL21
|
kelch-like 21 (Drosophila)
|
|
chr7_-_1498961
|
0.723
|
|
MICALL2
|
MICAL-like 2
|
|
chr3_+_154797644
|
0.714
|
NM_007287
|
MME
|
membrane metallo-endopeptidase
|
|
chr1_+_205198058
|
0.712
|
NM_001242925
|
TMCC2
|
transmembrane and coiled-coil domain family 2
|
|
chr1_+_156084541
|
0.672
|
|
LMNA
|
lamin A/C
|
|
chr11_-_65430427
|
0.658
|
NM_001145138
NM_001243984
NM_001243985
NM_021975
|
RELA
|
v-rel reticuloendotheliosis viral oncogene homolog A (avian)
|
|
chr1_-_6662898
|
0.655
|
NM_014851
|
KLHL21
|
kelch-like 21 (Drosophila)
|
|
chr16_+_57050985
|
0.636
|
NM_032206
|
NLRC5
|
NLR family, CARD domain containing 5
|
|
chr22_+_22723972
|
0.588
|
|
|
|
|
chr19_-_44174271
|
0.578
|
|
PLAUR
|
plasminogen activator, urokinase receptor
|
|
chr10_-_129691047
|
0.570
|
|
|
|
|
chr16_+_56642477
|
0.547
|
NM_005953
|
MT2A
|
metallothionein 2A
|
|
chr22_+_20861884
|
0.542
|
NM_001003891
NM_015889
|
MED15
|
mediator complex subunit 15
|
|
chr6_-_84140854
|
0.535
|
NM_002395
|
ME1
|
malic enzyme 1, NADP(+)-dependent, cytosolic
|
|
chr6_+_43597278
|
0.532
|
NM_001003690
|
MAD2L1BP
|
MAD2L1 binding protein
|
|
chr18_+_55888838
|
0.530
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr22_+_20861875
|
0.528
|
|
MED15
|
mediator complex subunit 15
|
|
chr22_+_20861886
|
0.528
|
|
MED15
|
mediator complex subunit 15
|
|
chr11_+_18415917
|
0.524
|
NM_001135239
NM_001165415
NM_001165416
NM_005566
|
LDHA
|
lactate dehydrogenase A
|
|
chr18_+_55888750
|
0.521
|
NM_001144966
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr7_-_115670828
|
0.521
|
NM_001018058
NM_012252
|
TFEC
|
transcription factor EC
|
|
chr16_+_30077120
|
0.519
|
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr18_+_55888799
|
0.518
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr16_+_30076993
|
0.508
|
NM_001243177
NM_184041
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr1_+_156084565
|
0.503
|
|
LMNA
|
lamin A/C
|
|
chr6_-_84140651
|
0.502
|
|
ME1
|
malic enzyme 1, NADP(+)-dependent, cytosolic
|
|
chr22_+_20861964
|
0.499
|
|
MED15
|
mediator complex subunit 15
|
|
chr21_+_43619798
|
0.498
|
NM_207627
NM_207628
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1
|
|
chr9_+_103191795
|
0.496
|
NM_001198806
|
C9orf30
|
chromosome 9 open reading frame 30
|
|
chr2_+_231191877
|
0.487
|
NM_138402
|
SP140L
|
SP140 nuclear body protein-like
|
|
chr19_+_41725135
|
0.477
|
|
AXL
|
AXL receptor tyrosine kinase
|
|
chr10_+_104155431
|
0.473
|
NM_002502
|
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100)
|
|
chr17_-_56595192
|
0.471
|
NM_004687
|
MTMR4
|
myotubularin related protein 4
|
|
chr16_+_30077146
|
0.467
|
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr6_-_138428477
|
0.466
|
NM_022121
|
PERP
|
PERP, TP53 apoptosis effector
|
|
chr22_+_20861898
|
0.465
|
|
MED15
|
mediator complex subunit 15
|
|
chr7_+_143078457
|
0.463
|
|
ZYX
|
zyxin
|
|
chr1_+_156084579
|
0.460
|
|
LMNA
|
lamin A/C
|
|
chr1_+_17866276
|
0.454
|
NM_018125
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like
|
|
chr11_-_65667860
|
0.448
|
NM_005438
|
FOSL1
|
FOS-like antigen 1
|
|
chr7_+_143078447
|
0.447
|
|
ZYX
|
zyxin
|
|
chr19_+_41725131
|
0.446
|
|
AXL
|
AXL receptor tyrosine kinase
|
|
chr22_+_39378404
|
0.441
|
NM_004900
|
APOBEC3D
APOBEC3B
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3D
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3B
|
|
chr1_+_40627066
|
0.437
|
|
RLF
|
rearranged L-myc fusion
|
|
chr7_+_143078444
|
0.434
|
|
ZYX
|
zyxin
|
|
chr18_+_61637262
|
0.434
|
NM_001031848
NM_002640
NM_198833
|
SERPINB8
|
serpin peptidase inhibitor, clade B (ovalbumin), member 8
|
|
chr10_-_129691202
|
0.432
|
NM_152311
|
CLRN3
|
clarin 3
|
|
chr1_-_89591685
|
0.432
|
NM_004120
|
GBP2
|
guanylate binding protein 2, interferon-inducible
|
|
chr7_+_143078395
|
0.431
|
|
ZYX
|
zyxin
|
|
chr10_-_95170
|
0.425
|
NM_177987
|
TUBB8
|
tubulin, beta 8 class VIII
|
|
chr17_+_74381419
|
0.419
|
|
SPHK1
|
sphingosine kinase 1
|
|
chr6_-_84140758
|
0.418
|
|
ME1
|
malic enzyme 1, NADP(+)-dependent, cytosolic
|
|
chr3_+_48507209
|
0.417
|
NM_016381
NM_033629
|
TREX1
|
three prime repair exonuclease 1
|
|
chr19_-_46285645
|
0.411
|
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr6_-_84140744
|
0.411
|
|
ME1
|
malic enzyme 1, NADP(+)-dependent, cytosolic
|
|
chr2_-_85641153
|
0.406
|
|
CAPG
|
capping protein (actin filament), gelsolin-like
|
|
chr7_+_143078399
|
0.400
|
|
ZYX
|
zyxin
|
|
chr21_+_30517897
|
0.394
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr3_+_48507621
|
0.393
|
|
TREX1
|
three prime repair exonuclease 1
|
|
chr1_+_36554452
|
0.390
|
NM_017825
|
ADPRHL2
|
ADP-ribosylhydrolase like 2
|
|
chr8_-_30515665
|
0.389
|
NM_002095
|
GTF2E2
|
general transcription factor IIE, polypeptide 2, beta 34kDa
|
|
chr17_+_74381186
|
0.389
|
NM_001142602
|
SPHK1
|
sphingosine kinase 1
|
|
chr20_-_52199635
|
0.387
|
NM_006526
|
ZNF217
|
zinc finger protein 217
|
|
chr6_-_134497069
|
0.381
|
NM_001143678
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr17_+_73717515
|
0.380
|
NM_000213
NM_001005731
|
ITGB4
|
integrin, beta 4
|
|
chr1_+_40627055
|
0.376
|
|
RLF
|
rearranged L-myc fusion
|
|
chr22_-_39151979
|
0.373
|
|
SUN2
|
Sad1 and UNC84 domain containing 2
|
|
chrX_-_129244422
|
0.372
|
|
ELF4
|
E74-like factor 4 (ets domain transcription factor)
|
|
chrX_-_48937560
|
0.369
|
NM_001029896
|
WDR45
|
WD repeat domain 45
|
|
chr19_+_58695199
|
0.366
|
|
ZNF274
|
zinc finger protein 274
|
|
chr7_+_143078358
|
0.362
|
NM_001010972
NM_003461
|
ZYX
|
zyxin
|
|
chrX_-_48937457
|
0.360
|
|
WDR45
|
WD repeat domain 45
|
|
chr10_+_134901408
|
0.360
|
NM_001083909
|
GPR123
|
G protein-coupled receptor 123
|
|
chr1_-_27816676
|
0.355
|
NM_001201404
NM_006990
|
WASF2
|
WAS protein family, member 2
|
|
chr12_-_51403028
|
0.352
|
NM_001174130
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2
|
|
chr1_-_27816542
|
0.350
|
|
WASF2
|
WAS protein family, member 2
|
|
chr1_+_36621457
|
0.347
|
|
MAP7D1
|
MAP7 domain containing 1
|
|
chrX_-_129244471
|
0.345
|
NM_001421
|
ELF4
|
E74-like factor 4 (ets domain transcription factor)
|
|
chr22_-_39152008
|
0.344
|
NM_001199579
NM_015374
|
SUN2
|
Sad1 and UNC84 domain containing 2
|
|
chr1_+_36621751
|
0.341
|
NM_018067
|
MAP7D1
|
MAP7 domain containing 1
|
|
chr18_-_25756946
|
0.337
|
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal)
|
|
chr1_-_109969044
|
0.335
|
|
PSMA5
|
proteasome (prosome, macropain) subunit, alpha type, 5
|
|
chr1_+_155853546
|
0.334
|
|
SYT11
|
synaptotagmin XI
|
|
chr22_-_39151457
|
0.333
|
NM_001199580
|
SUN2
|
Sad1 and UNC84 domain containing 2
|
|
chr1_-_27816667
|
0.330
|
|
WASF2
|
WAS protein family, member 2
|
|
chr5_+_70944968
|
0.328
|
|
MCCC2
|
methylcrotonoyl-CoA carboxylase 2 (beta)
|
|
chr17_+_18380097
|
0.327
|
NM_001040078
|
LGALS9B
LGALS9C
|
lectin, galactoside-binding, soluble, 9B
lectin, galactoside-binding, soluble, 9C
|
|
chr16_-_1429612
|
0.327
|
NM_001193389
NM_023076
|
UNKL
|
unkempt homolog (Drosophila)-like
|
|
chr19_-_43969723
|
0.323
|
NM_014400
|
LYPD3
|
LY6/PLAUR domain containing 3
|
|
chr2_-_73316782
|
0.323
|
|
RAB11FIP5
|
RAB11 family interacting protein 5 (class I)
|
|
chr3_+_11178778
|
0.321
|
NM_001098213
|
HRH1
|
histamine receptor H1
|
|
chr1_-_109968951
|
0.315
|
|
PSMA5
|
proteasome (prosome, macropain) subunit, alpha type, 5
|
|
chr22_-_30642683
|
0.315
|
NM_002309
|
LIF
|
leukemia inhibitory factor (cholinergic differentiation factor)
|
|
chr16_+_67311412
|
0.310
|
NM_015432
|
PLEKHG4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4
|
|
chr7_+_143058794
|
0.309
|
|
ZYX
|
zyxin
|
|
chr12_-_56236614
|
0.308
|
NM_002429
|
MMP19
|
matrix metallopeptidase 19
|
|
chr17_+_21187967
|
0.305
|
NM_145109
|
MAP2K3
|
mitogen-activated protein kinase kinase 3
|
|
chr3_-_151034514
|
0.303
|
NM_023915
|
GPR87
|
G protein-coupled receptor 87
|
|
chr6_+_29910308
|
0.302
|
|
HLA-A
|
major histocompatibility complex, class I, A
|
|
chr1_+_244515936
|
0.300
|
NM_001012970
|
C1orf100
|
chromosome 1 open reading frame 100
|
|
chr16_+_56685810
|
0.296
|
NM_005947
|
MT1B
|
metallothionein 1B
|
|
chr6_-_35888816
|
0.295
|
|
SRPK1
|
SRSF protein kinase 1
|
|
chr17_-_55980749
|
0.292
|
NM_017949
|
CUEDC1
|
CUE domain containing 1
|
|
chr1_-_109969058
|
0.291
|
NM_001199772
NM_002790
NM_001199773
NM_001199774
|
PSMA5
|
proteasome (prosome, macropain) subunit, alpha type, 5
|
|
chr21_+_42733916
|
0.290
|
NM_002463
|
LOC100131190
MX2
|
uncharacterized LOC100131190
myxovirus (influenza virus) resistance 2 (mouse)
|
|
chr6_+_138483024
|
0.289
|
NM_020340
|
KIAA1244
|
KIAA1244
|
|
chr11_-_65430214
|
0.287
|
|
RELA
|
v-rel reticuloendotheliosis viral oncogene homolog A (avian)
|
|
chr17_+_74380943
|
0.282
|
|
SPHK1
|
sphingosine kinase 1
|
|
chr2_+_69201704
|
0.278
|
NM_019617
|
GKN1
|
gastrokine 1
|
|
chr12_-_76478737
|
0.275
|
NM_004537
NM_139207
|
NAP1L1
|
nucleosome assembly protein 1-like 1
|
|
chr18_-_74207145
|
0.273
|
NM_014643
|
ZNF516
|
zinc finger protein 516
|
|
chr9_+_99691285
|
0.273
|
NM_001045477
NM_001170741
|
FAM22G
|
family with sequence similarity 22, member G
|
|
chr21_-_36421586
|
0.273
|
NM_001754
|
RUNX1
|
runt-related transcription factor 1
|
|
chr17_-_33390676
|
0.272
|
NM_001017368
|
RFFL
|
ring finger and FYVE-like domain containing 1
|
|
chrX_-_129244652
|
0.272
|
NM_001127197
|
ELF4
|
E74-like factor 4 (ets domain transcription factor)
|
|
chr17_+_21188032
|
0.271
|
|
MAP2K3
|
mitogen-activated protein kinase kinase 3
|
|
chr1_+_40627031
|
0.271
|
NM_012421
|
RLF
|
rearranged L-myc fusion
|
|
chr1_-_45308603
|
0.271
|
NM_001166292
NM_003738
|
PTCH2
|
patched 2
|
|
chr10_-_35363167
|
0.266
|
NM_001198778
NM_001198779
|
CUL2
|
cullin 2
|
|
chr1_+_19638739
|
0.265
|
NM_001040125
NM_001040126
NM_017765
|
PQLC2
|
PQ loop repeat containing 2
|
|
chr14_-_24777064
|
0.265
|
|
|
|
|
chr6_-_119031230
|
0.263
|
NM_001178035
|
CEP85L
|
centrosomal protein 85kDa-like
|
|
chr12_+_64845839
|
0.262
|
NM_013254
|
TBK1
|
TANK-binding kinase 1
|
|
chr21_-_36421578
|
0.261
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr10_+_104154228
|
0.257
|
NM_001077493
NM_001077494
|
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100)
|
|
chr16_+_67312057
|
0.257
|
NM_001129727
|
PLEKHG4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4
|
|
chr17_+_60447578
|
0.257
|
NM_001144933
|
EFCAB3
|
EF-hand calcium binding domain 3
|
|
chr22_-_30942668
|
0.256
|
NM_001193336
|
SEC14L6
|
SEC14-like 6 (S. cerevisiae)
|
|
chr3_-_150480917
|
0.254
|
|
SIAH2
|
seven in absentia homolog 2 (Drosophila)
|
|
chr10_+_121410751
|
0.254
|
NM_004281
|
BAG3
|
BCL2-associated athanogene 3
|
|
chr6_-_74161824
|
0.252
|
|
MB21D1
|
Mab-21 domain containing 1
|
|
chr8_-_125579395
|
0.247
|
|
MTSS1
|
metastasis suppressor 1
|
|
chr7_-_150721551
|
0.247
|
NM_173681
|
ATG9B
|
ATG9 autophagy related 9 homolog B (S. cerevisiae)
|
|
chrX_+_133371076
|
0.246
|
NM_001101357
|
CCDC160
|
coiled-coil domain containing 160
|
|
chr15_+_22892714
|
0.245
|
|
CYFIP1
|
cytoplasmic FMR1 interacting protein 1
|
|
chr2_+_207630310
|
0.243
|
|
FASTKD2
|
FAST kinase domains 2
|
|
chr11_-_92931045
|
0.242
|
NM_152313
|
SLC36A4
|
solute carrier family 36 (proton/amino acid symporter), member 4
|
|
chr17_+_21191347
|
0.242
|
NM_002756
|
MAP2K3
|
mitogen-activated protein kinase kinase 3
|
|
chr19_-_6591012
|
0.242
|
|
CD70
|
CD70 molecule
|
|
chr6_-_30654978
|
0.242
|
|
PPP1R18
|
protein phosphatase 1, regulatory subunit 18
|
|
chr5_+_40841296
|
0.241
|
NM_032587
|
CARD6
|
caspase recruitment domain family, member 6
|
|
chr5_-_75919051
|
0.239
|
NM_004101
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2
|
|
chr16_-_84538219
|
0.238
|
NM_020947
|
KIAA1609
|
KIAA1609
|
|
chr5_+_42756919
|
0.237
|
NM_001134848
|
CCDC152
|
coiled-coil domain containing 152
|
|
chr1_-_158656487
|
0.236
|
NM_003126
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2)
|
|
chr15_+_22892517
|
0.235
|
|
CYFIP1
|
cytoplasmic FMR1 interacting protein 1
|
|
chr12_-_323255
|
0.234
|
NM_001122848
NM_001206931
|
SLC6A12
|
solute carrier family 6 (neurotransmitter transporter, betaine/GABA), member 12
|
|
chr16_-_28607694
|
0.234
|
NM_177528
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2
|
|
chr16_+_89988416
|
0.234
|
NM_001197181
|
TUBB3
|
tubulin, beta 3 class III
|
|
chr17_-_28257017
|
0.234
|
NM_033389
|
SSH2
|
slingshot homolog 2 (Drosophila)
|
|
chr15_+_22892683
|
0.232
|
NM_014608
|
CYFIP1
|
cytoplasmic FMR1 interacting protein 1
|
|
chr3_-_150480963
|
0.230
|
NM_005067
|
SIAH2
|
seven in absentia homolog 2 (Drosophila)
|
|
chr22_+_22676814
|
0.227
|
|
|
|
|
chr4_-_100356290
|
0.227
|
NM_001166504
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide
|
|
chr1_+_15480213
|
0.226
|
NM_001136218
NM_018022
|
TMEM51
|
transmembrane protein 51
|
|
chr16_-_12010518
|
0.225
|
NM_001130007
|
GSPT1
|
G1 to S phase transition 1
|
|
chr3_+_184016985
|
0.225
|
NM_002808
|
PSMD2
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 2
|
|
chr1_+_223889253
|
0.224
|
NM_001146068
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr12_-_58165877
|
0.223
|
NM_005371
NM_023033
|
METTL1
|
methyltransferase like 1
|
|
chr10_+_71075609
|
0.221
|
NM_033496
|
HK1
|
hexokinase 1
|
|
chr15_+_22892723
|
0.220
|
|
CYFIP1
|
cytoplasmic FMR1 interacting protein 1
|
|
chr1_-_108231122
|
0.220
|
NM_001079874
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr1_-_1149495
|
0.218
|
NM_003327
|
TNFRSF4
|
tumor necrosis factor receptor superfamily, member 4
|