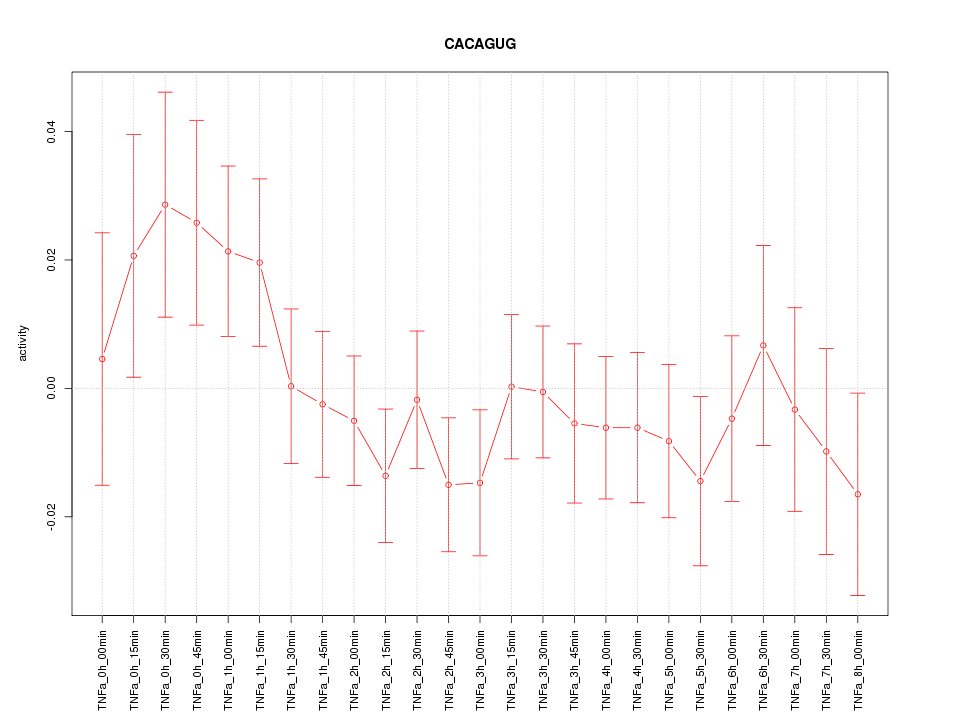
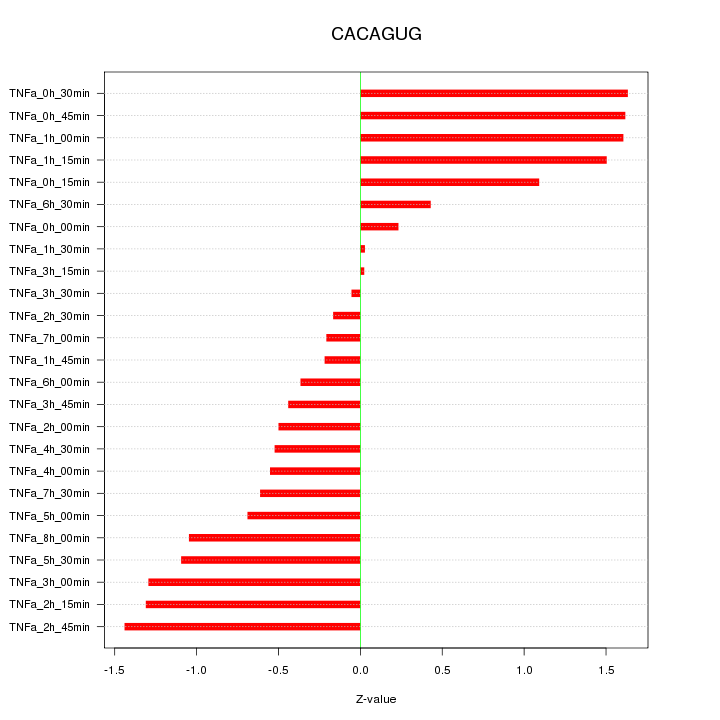
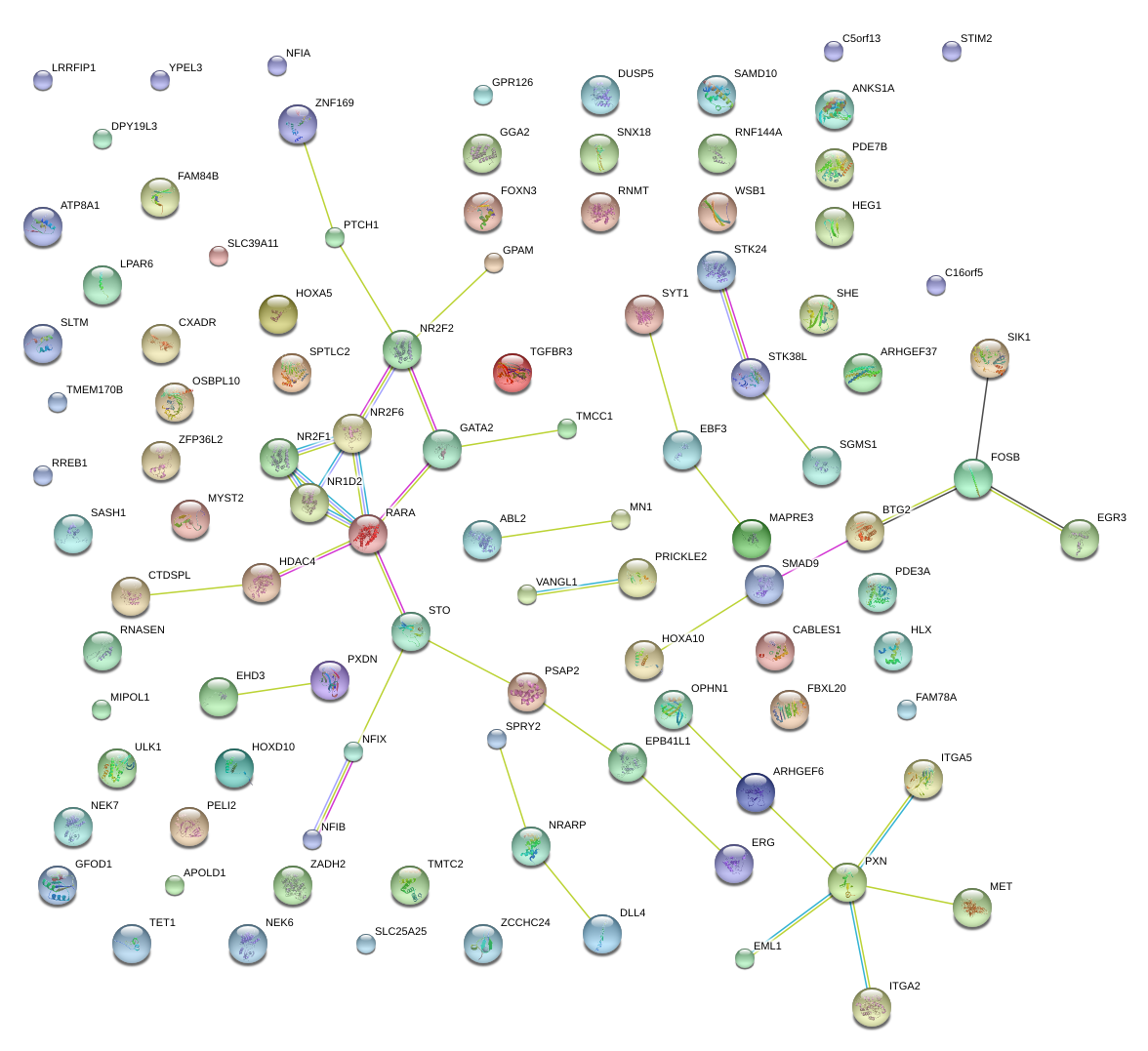
|
chr8_-_22549901
|
4.574
|
NM_001199880
|
EGR3
|
early growth response 3
|
|
chr8_-_22550708
|
4.519
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr8_-_22550057
|
4.476
|
NM_001199881
|
EGR3
|
early growth response 3
|
|
chr2_+_7057471
|
3.428
|
NM_014746
|
RNF144A
|
ring finger protein 144A
|
|
chr9_-_134151905
|
3.387
|
NM_033387
|
FAM78A
|
family with sequence similarity 78, member A
|
|
chr6_+_136172802
|
3.151
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr6_-_13408368
|
3.097
|
NM_001242630
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1
|
|
chr1_+_221052735
|
2.958
|
NM_021958
|
HLX
|
H2.0-like homeobox
|
|
chr1_-_92371558
|
2.932
|
NM_001195684
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr1_-_92351613
|
2.798
|
NM_001195683
NM_003243
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr19_+_45971249
|
2.703
|
NM_001114171
NM_006732
|
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B
|
|
chr10_-_131762017
|
2.600
|
NM_001005463
|
EBF3
|
early B-cell factor 3
|
|
chr8_-_127570710
|
2.537
|
NM_174911
|
FAM84B
|
family with sequence similarity 84, member B
|
|
chr14_+_56584854
|
2.530
|
NM_021255
|
PELI2
|
pellino homolog 2 (Drosophila)
|
|
chr7_-_27183225
|
2.482
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr18_+_20715726
|
2.341
|
NM_001100619
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1
|
|
chr18_+_20735788
|
2.324
|
NM_138375
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1
|
|
chr1_-_154474524
|
2.310
|
NM_001010846
|
SHE
|
Src homology 2 domain containing E
|
|
chr10_-_81205091
|
2.306
|
NM_153367
|
ZCCHC24
|
zinc finger, CCHC domain containing 24
|
|
chr22_-_28197469
|
2.250
|
NM_002430
|
MN1
|
meningioma (disrupted in balanced translocation) 1
|
|
chr6_+_142622797
|
2.249
|
NM_001032394
NM_001032395
NM_020455
NM_198569
|
GPR126
|
G protein-coupled receptor 126
|
|
chr7_-_28998028
|
2.163
|
NM_014817
|
TRIL
|
TLR4 interactor with leucine-rich repeats
|
|
chr9_-_140196702
|
2.068
|
NM_001004354
|
NRARP
|
NOTCH-regulated ankyrin repeat protein
|
|
chr1_+_61542928
|
2.052
|
NM_001145511
|
NFIA
|
nuclear factor I/A
|
|
chr7_-_27213873
|
2.039
|
NM_018951
|
HOXA10
|
homeobox A10
|
|
chr1_+_61547625
|
2.030
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr13_-_37494370
|
1.924
|
NM_001127217
NM_005905
|
SMAD9
|
SMAD family member 9
|
|
chr9_+_127054246
|
1.904
|
NM_001166170
NM_001166171
|
NEK6
|
NIMA (never in mitosis gene a)-related kinase 6
|
|
chr21_-_44846927
|
1.863
|
NM_173354
|
SIK1
|
salt-inducible kinase 1
|
|
chr22_+_51113069
|
1.846
|
NM_001080420
|
SHANK3
|
SH3 and multiple ankyrin repeat domains 3
|
|
chr1_+_61547388
|
1.845
|
NM_001145512
|
NFIA
|
nuclear factor I/A
|
|
chr3_+_37903643
|
1.820
|
NM_001008392
NM_005808
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like
|
|
chr13_-_49001042
|
1.771
|
NM_001162497
|
LPAR6
|
lysophosphatidic acid receptor 6
|
|
chr15_+_41221530
|
1.759
|
NM_019074
|
DLL4
|
delta-like 4 (Drosophila)
|
|
chr5_-_111093251
|
1.736
|
NM_001142479
NM_001142480
NM_001142478
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr14_+_100259445
|
1.736
|
NM_001008707
NM_004434
|
EML1
|
echinoderm microtubule associated protein like 1
|
|
chr9_+_127054567
|
1.726
|
NM_001166169
|
NEK6
|
NIMA (never in mitosis gene a)-related kinase 6
|
|
chr1_+_203274646
|
1.699
|
NM_006763
|
BTG2
|
BTG family, member 2
|
|
chr1_+_116185249
|
1.622
|
NM_001172412
|
VANGL1
|
vang-like 1 (van gogh, Drosophila)
|
|
chr9_+_127019782
|
1.615
|
NM_001166167
NM_001145001
NM_014397
|
NEK6
|
NIMA (never in mitosis gene a)-related kinase 6
|
|
chr5_-_111093030
|
1.594
|
NM_001142482
NM_001142481
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr6_+_11538486
|
1.575
|
NM_001100829
|
TMEM170B
|
transmembrane protein 170B
|
|
chr1_+_116184573
|
1.564
|
NM_001172411
NM_138959
|
VANGL1
|
vang-like 1 (van gogh, Drosophila)
|
|
chr5_-_111093792
|
1.560
|
NM_001142477
NM_001142476
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_+_52284903
|
1.559
|
NM_002203
|
ITGA2
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor)
|
|
chr9_+_127020462
|
1.559
|
NM_001166168
|
NEK6
|
NIMA (never in mitosis gene a)-related kinase 6
|
|
chr5_-_111312522
|
1.490
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_-_111092918
|
1.475
|
NM_004772
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr13_-_48987652
|
1.462
|
NM_001162498
|
LPAR6
|
lysophosphatidic acid receptor 6
|
|
chr21_+_18885104
|
1.444
|
NM_001207063
NM_001207064
NM_001207065
NM_001207066
NM_001338
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chr7_+_116312412
|
1.369
|
NM_000245
NM_001127500
|
MET
|
met proto-oncogene (hepatocyte growth factor receptor)
|
|
chr3_-_128212027
|
1.335
|
NM_032638
|
GATA2
|
GATA binding protein 2
|
|
chr2_+_31456879
|
1.297
|
NM_014600
|
EHD3
|
EH-domain containing 3
|
|
chr12_+_12938540
|
1.281
|
NM_030817
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr12_+_20522178
|
1.280
|
NM_000921
|
PDE3A
|
phosphodiesterase 3A, cGMP-inhibited
|
|
chr20_+_34742656
|
1.271
|
NM_012156
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr12_+_79439432
|
1.268
|
NM_001135806
|
SYT1
|
synaptotagmin I
|
|
chr10_-_52383611
|
1.254
|
NM_147156
|
SGMS1
|
sphingomyelin synthase 1
|
|
chr12_+_79257772
|
1.253
|
NM_001135805
|
SYT1
|
synaptotagmin I
|
|
chr5_-_111091947
|
1.249
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr3_-_128207366
|
1.243
|
NM_001145661
|
GATA2
|
GATA binding protein 2
|
|
chr2_-_43453737
|
1.203
|
NM_006887
|
ZFP36L2
|
zinc finger protein 36, C3H type-like 2
|
|
chr12_+_27397077
|
1.198
|
NM_015000
|
STK38L
|
serine/threonine kinase 38 like
|
|
chr5_+_176560056
|
1.195
|
NM_172349
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr6_-_13486382
|
1.182
|
NM_001242628
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1
|
|
chr14_-_89883306
|
1.180
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr3_-_32023237
|
1.165
|
NM_001174060
NM_017784
|
OSBPL10
|
oxysterol binding protein-like 10
|
|
chr17_+_25621105
|
1.156
|
NM_015626
NM_134265
|
WSB1
|
WD repeat and SOCS box containing 1
|
|
chr12_+_79258448
|
1.149
|
NM_005639
|
SYT1
|
synaptotagmin I
|
|
chr5_-_31532164
|
1.146
|
NM_001100412
NM_013235
|
DROSHA
|
drosha, ribonuclease type III
|
|
chr12_-_120687956
|
1.145
|
NM_025157
|
PXN
|
paxillin
|
|
chrX_-_135863502
|
1.143
|
NM_004840
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr12_+_12878718
|
1.094
|
NM_001130415
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr9_-_14398981
|
1.092
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chr19_+_32897021
|
1.084
|
NM_207325
|
DPY19L3
|
dpy-19-like 3 (C. elegans)
|
|
chr10_+_112257624
|
1.083
|
NM_004419
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr19_-_17356022
|
1.079
|
NM_005234
|
NR2F6
|
nuclear receptor subfamily 2, group F, member 6
|
|
chr20_+_34700257
|
1.060
|
NM_177996
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr19_+_32896515
|
1.045
|
NM_001172774
|
DPY19L3
|
dpy-19-like 3 (C. elegans)
|
|
chr2_+_238536183
|
1.042
|
NM_001137550
|
LRRFIP1
|
leucine rich repeat (in FLII) interacting protein 1
|
|
chr16_-_30107492
|
1.032
|
NM_031477
NM_001145524
|
YPEL3
|
yippee-like 3 (Drosophila)
|
|
chr3_-_124774735
|
1.027
|
NM_020733
|
HEG1
|
HEG homolog 1 (zebrafish)
|
|
chr18_-_72921280
|
1.025
|
NM_175907
|
ZADH2
|
zinc binding alcohol dehydrogenase domain containing 2
|
|
chr17_+_47865980
|
1.018
|
NM_001199155
NM_001199156
NM_001199157
NM_001199158
NM_007067
|
KAT7
|
K(lysine) acetyltransferase 7
|
|
chr5_+_53813494
|
1.013
|
NM_001145427
NM_001102575
NM_052870
|
SNX18
|
sorting nexin 18
|
|
chr12_-_120703549
|
1.011
|
NM_001080855
NM_001243756
NM_002859
|
PXN
|
paxillin
|
|
chr3_-_129599220
|
1.007
|
NM_001017395
|
TMCC1
|
transmembrane and coiled-coil domain family 1
|
|
chr17_-_37557851
|
0.995
|
NM_001184906
NM_032875
|
FBXL20
|
F-box and leucine-rich repeat protein 20
|
|
chr6_+_34857021
|
0.992
|
NM_015245
|
ANKS1A
|
ankyrin repeat and sterile alpha motif domain containing 1A
|
|
chr20_-_62610990
|
0.987
|
NM_080621
|
SAMD10
|
sterile alpha motif domain containing 10
|
|
chr9_-_14314036
|
0.984
|
NM_001190737
NM_005596
|
NFIB
|
nuclear factor I/B
|
|
chr3_-_129612371
|
0.983
|
NM_001128224
|
TMCC1
|
transmembrane and coiled-coil domain family 1
|
|
chr14_-_90085325
|
0.976
|
NM_001085471
|
FOXN3
|
forkhead box N3
|
|
chr12_+_83080901
|
0.973
|
NM_152588
|
TMTC2
|
transmembrane and tetratricopeptide repeat containing 2
|
|
chr6_+_148663728
|
0.925
|
NM_015278
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr17_-_71088832
|
0.913
|
NM_001159770
NM_139177
|
SLC39A11
|
solute carrier family 39 (metal ion transporter), member 11
|
|
chr16_-_23521803
|
0.896
|
NM_015044
|
GGA2
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 2
|
|
chr14_-_78083109
|
0.893
|
NM_004863
|
SPTLC2
|
serine palmitoyltransferase, long chain base subunit 2
|
|
chr15_+_96876568
|
0.884
|
NM_001145157
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr4_-_42658883
|
0.875
|
NM_001105529
NM_006095
|
ATP8A1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1
|
|
chr5_+_148961035
|
0.874
|
NM_001001669
|
ARHGEF37
|
Rho guanine nucleotide exchange factor (GEF) 37
|
|
chr16_-_4588379
|
0.869
|
NM_001199054
|
C16orf5
|
chromosome 16 open reading frame 5
|
|
chr3_+_23987514
|
0.859
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr4_+_26862312
|
0.849
|
NM_001169117
NM_001169118
NM_020860
|
STIM2
|
stromal interaction molecule 2
|
|
chr1_-_179112178
|
0.835
|
NM_001136000
NM_001168239
NM_005158
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr14_+_37667116
|
0.816
|
NM_001195296
NM_001195297
NM_138731
|
MIPOL1
|
mirror-image polydactyly 1
|
|
chr21_-_40033617
|
0.811
|
NM_001243432
NM_001136154
NM_004449
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr15_+_96875793
|
0.798
|
NM_001145156
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr12_+_132379262
|
0.788
|
NM_003565
|
ULK1
|
unc-51-like kinase 1 (C. elegans)
|
|
chr3_-_64211111
|
0.787
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr13_-_99174251
|
0.772
|
NM_003576
|
STK24
|
serine/threonine kinase 24
|
|
chr9_-_98269480
|
0.765
|
NM_001083607
|
PTCH1
|
patched 1
|
|
chr9_+_130830447
|
0.752
|
NM_001006641
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr9_+_130860816
|
0.737
|
NM_052901
NM_001006643
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr21_-_39870303
|
0.735
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr13_-_99229226
|
0.734
|
NM_001032296
|
STK24
|
serine/threonine kinase 24
|
|
chr13_-_80915041
|
0.727
|
NM_005842
|
SPRY2
|
sprouty homolog 2 (Drosophila)
|
|
chrX_-_67653169
|
0.725
|
NM_002547
|
OPHN1
|
oligophrenin 1
|
|
chr12_-_54813010
|
0.715
|
NM_002205
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr6_-_13487772
|
0.709
|
NM_001242629
NM_018988
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1
|
|
chr9_-_98270830
|
0.708
|
NM_000264
|
PTCH1
|
patched 1
|
|
chr6_+_7107986
|
0.707
|
NM_001003698
NM_001003699
NM_001003700
|
RREB1
|
ras responsive element binding protein 1
|
|
chr9_+_130854127
|
0.706
|
NM_001006642
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr15_+_96869156
|
0.703
|
NM_001145155
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr9_-_98270321
|
0.698
|
NM_001083604
NM_001083605
|
PTCH1
|
patched 1
|
|
chr17_+_38474472
|
0.697
|
NM_001145301
NM_001145302
|
RARA
|
retinoic acid receptor, alpha
|
|
chr10_+_70320044
|
0.694
|
NM_030625
|
TET1
|
tet methylcytosine dioxygenase 1
|
|
chr2_-_240322620
|
0.683
|
NM_006037
|
HDAC4
|
histone deacetylase 4
|
|
chr10_-_113943467
|
0.681
|
NM_001244949
NM_020918
|
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial
|
|
chr3_+_15247727
|
0.668
|
NM_014296
|
CAPN7
|
calpain 7
|
|
chr15_+_47476225
|
0.659
|
NM_001198999
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr14_-_39901619
|
0.653
|
NM_203301
|
FBXO33
|
F-box protein 33
|
|
chr6_+_41606193
|
0.649
|
NM_005586
|
MDFI
|
MyoD family inhibitor
|
|
chr3_-_141719188
|
0.645
|
NM_001178140
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr12_+_32112286
|
0.638
|
NM_018169
|
C12orf35
|
chromosome 12 open reading frame 35
|
|
chr12_-_118406027
|
0.636
|
NM_173598
|
KSR2
|
kinase suppressor of ras 2
|
|
chr8_-_57123858
|
0.632
|
NM_001114634
NM_001114635
NM_002655
|
PLAG1
|
pleiomorphic adenoma gene 1
|
|
chr3_-_13114547
|
0.629
|
NM_001134382
|
IQSEC1
|
IQ motif and Sec7 domain 1
|
|
chr1_-_225840660
|
0.629
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr14_-_91526912
|
0.618
|
NM_004755
NM_182398
|
RPS6KA5
|
ribosomal protein S6 kinase, 90kDa, polypeptide 5
|
|
chr1_+_76540374
|
0.616
|
NM_001160011
NM_152996
|
ST6GALNAC3
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3
|
|
chr15_+_96873845
|
0.614
|
NM_021005
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr1_-_33841142
|
0.614
|
NM_198040
|
PHC2
|
polyhomeotic homolog 2 (Drosophila)
|
|
chr6_+_7107799
|
0.612
|
NM_001168344
|
RREB1
|
ras responsive element binding protein 1
|
|
chr11_+_33563772
|
0.611
|
NM_012194
|
C11orf41
|
chromosome 11 open reading frame 41
|
|
chr16_-_70719854
|
0.608
|
NM_138383
|
MTSS1L
|
metastasis suppressor 1-like
|
|
chr12_-_31743822
|
0.608
|
NM_144973
|
DENND5B
|
DENN/MADD domain containing 5B
|
|
chr13_-_41240607
|
0.606
|
NM_002015
|
FOXO1
|
forkhead box O1
|
|
chr13_+_33160537
|
0.598
|
NM_015032
|
PDS5B
|
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae)
|
|
chr4_-_48782281
|
0.596
|
NM_015030
|
FRYL
|
FRY-like
|
|
chr1_-_179198814
|
0.595
|
NM_001136001
NM_001168236
NM_001168237
NM_001168238
NM_007314
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr5_-_93447237
|
0.595
|
NM_001163417
NM_001163418
NM_032042
|
FAM172A
|
family with sequence similarity 172, member A
|
|
chr16_-_4588738
|
0.591
|
NM_001199055
NM_001199056
NM_013399
|
C16orf5
|
chromosome 16 open reading frame 5
|
|
chr3_-_39195101
|
0.585
|
NM_033027
|
CSRNP1
|
cysteine-serine-rich nuclear protein 1
|
|
chr1_-_68299047
|
0.584
|
NM_018841
|
GNG12
|
guanine nucleotide binding protein (G protein), gamma 12
|
|
chr17_+_38465422
|
0.582
|
NM_000964
|
RARA
|
retinoic acid receptor, alpha
|
|
chr8_+_79428335
|
0.576
|
NM_006823
NM_181839
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha
|
|
chr2_+_203241044
|
0.562
|
NM_001204
|
BMPR2
|
bone morphogenetic protein receptor, type II (serine/threonine kinase)
|
|
chr6_-_119399759
|
0.562
|
NM_024581
|
FAM184A
|
family with sequence similarity 184, member A
|
|
chr17_+_61086897
|
0.546
|
NM_025185
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2
|
|
chr9_+_129089092
|
0.543
|
NM_001011703
NM_033446
|
FAM125B
|
family with sequence similarity 125, member B
|
|
chr17_+_38278789
|
0.535
|
NM_001012241
|
MSL1
|
male-specific lethal 1 homolog (Drosophila)
|
|
chr9_-_138987096
|
0.532
|
NM_144653
|
NACC2
|
NACC family member 2, BEN and BTB (POZ) domain containing
|
|
chr12_-_12419702
|
0.527
|
NM_002336
|
LRP6
|
low density lipoprotein receptor-related protein 6
|
|
chr10_-_65225605
|
0.527
|
NM_032776
|
JMJD1C
|
jumonji domain containing 1C
|
|
chr8_-_81787015
|
0.517
|
NM_001033723
|
ZNF704
|
zinc finger protein 704
|
|
chr4_-_7941092
|
0.516
|
NM_001134647
NM_198595
|
AFAP1
|
actin filament associated protein 1
|
|
chr17_-_58603492
|
0.510
|
NM_006380
|
APPBP2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2
|
|
chr14_-_30396811
|
0.508
|
NM_002742
|
PRKD1
|
protein kinase D1
|
|
chr6_-_33267128
|
0.508
|
NM_001243738
NM_004761
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2
|
|
chr17_-_35969401
|
0.498
|
NM_001163544
NM_001163545
NM_001163546
NM_001163547
NM_007247
NM_080550
NM_198882
|
SYNRG
|
synergin, gamma
|
|
chr15_+_91447419
|
0.497
|
NM_006122
|
MAN2A2
|
mannosidase, alpha, class 2A, member 2
|
|
chr1_-_8877379
|
0.496
|
NM_001042681
NM_012102
|
RERE
|
arginine-glutamic acid dipeptide (RE) repeats
|
|
chr3_+_23986735
|
0.494
|
NM_005126
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr10_+_22610138
|
0.486
|
NM_005180
|
BMI1
|
BMI1 polycomb ring finger oncogene
|
|
chr3_-_141868208
|
0.485
|
NM_001178138
NM_001178139
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr11_+_121322848
|
0.484
|
NM_003105
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chr11_+_118230292
|
0.483
|
NM_001204077
NM_004788
|
UBE4A
|
ubiquitination factor E4A
|
|
chr4_-_170192193
|
0.481
|
NM_020870
|
SH3RF1
|
SH3 domain containing ring finger 1
|
|
chr1_-_8483701
|
0.478
|
NM_001042682
|
RERE
|
arginine-glutamic acid dipeptide (RE) repeats
|
|
chr9_-_98269686
|
0.476
|
NM_001083606
|
PTCH1
|
patched 1
|
|
chr2_+_242641310
|
0.474
|
NM_032329
|
ING5
|
inhibitor of growth family, member 5
|
|
chr17_-_49337370
|
0.471
|
NM_017643
|
MBTD1
|
mbt domain containing 1
|
|
chr12_-_109125289
|
0.470
|
NM_014325
|
CORO1C
|
coronin, actin binding protein, 1C
|
|
chr22_-_37098620
|
0.469
|
NM_006078
|
CACNG2
|
calcium channel, voltage-dependent, gamma subunit 2
|
|
chr15_-_65067696
|
0.465
|
NM_194272
|
RBPMS2
|
RNA binding protein with multiple splicing 2
|
|
chr5_+_176560627
|
0.463
|
NM_022455
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr11_-_88781039
|
0.461
|
NM_001143831
|
GRM5
|
glutamate receptor, metabotropic 5
|
|
chr17_-_46692300
|
0.459
|
NM_024016
|
HOXB8
|
homeobox B8
|
|
chr17_-_8534008
|
0.458
|
NM_005964
|
MYH10
|
myosin, heavy chain 10, non-muscle
|
|
chr12_-_71031174
|
0.455
|
NM_001109754
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B
|
|
chr6_-_119470336
|
0.453
|
NM_001100411
|
FAM184A
|
family with sequence similarity 184, member A
|
|
chr15_+_76352270
|
0.452
|
NM_152335
|
C15orf27
|
chromosome 15 open reading frame 27
|
|
chr22_-_29075708
|
0.447
|
NM_001145418
|
TTC28
|
tetratricopeptide repeat domain 28
|
|
chr5_+_78531848
|
0.446
|
NM_152405
|
JMY
|
junction mediating and regulatory protein, p53 cofactor
|
|
chr10_+_35535952
|
0.446
|
NM_181698
|
CCNY
|
cyclin Y
|
|
chr21_+_35445808
|
0.445
|
NM_032476
NM_006933
|
MRPS6
SLC5A3
|
mitochondrial ribosomal protein S6
solute carrier family 5 (sodium/myo-inositol cotransporter), member 3
|
|
chr8_-_103424897
|
0.442
|
NM_015902
|
UBR5
|
ubiquitin protein ligase E3 component n-recognin 5
|
|
chr3_+_50654558
|
0.440
|
NM_001243925
NM_004635
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3
|
|
chr6_-_8064642
|
0.440
|
NM_001199322
NM_001199323
NM_201280
|
MUTED
|
muted homolog (mouse)
|