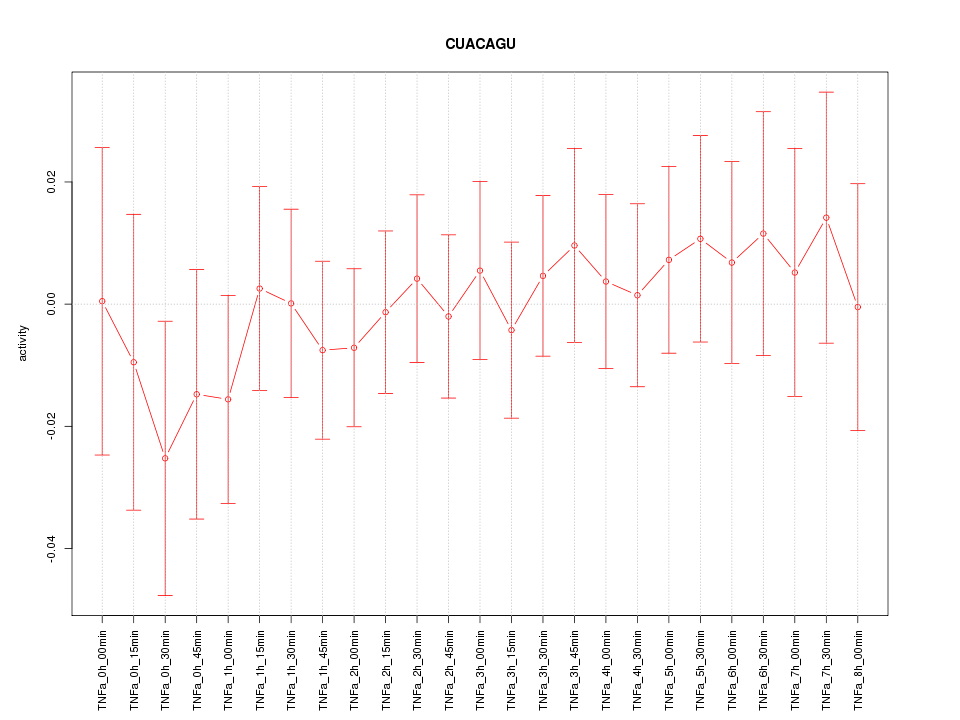
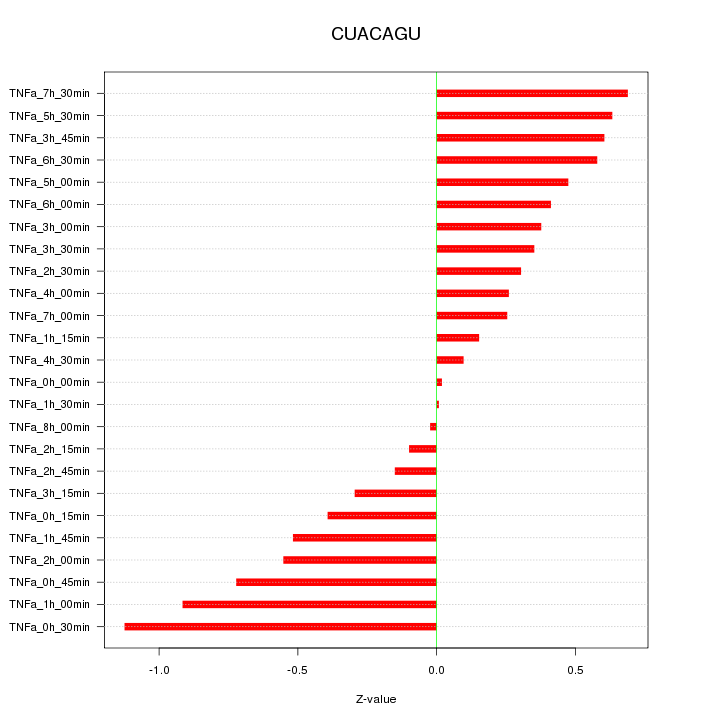
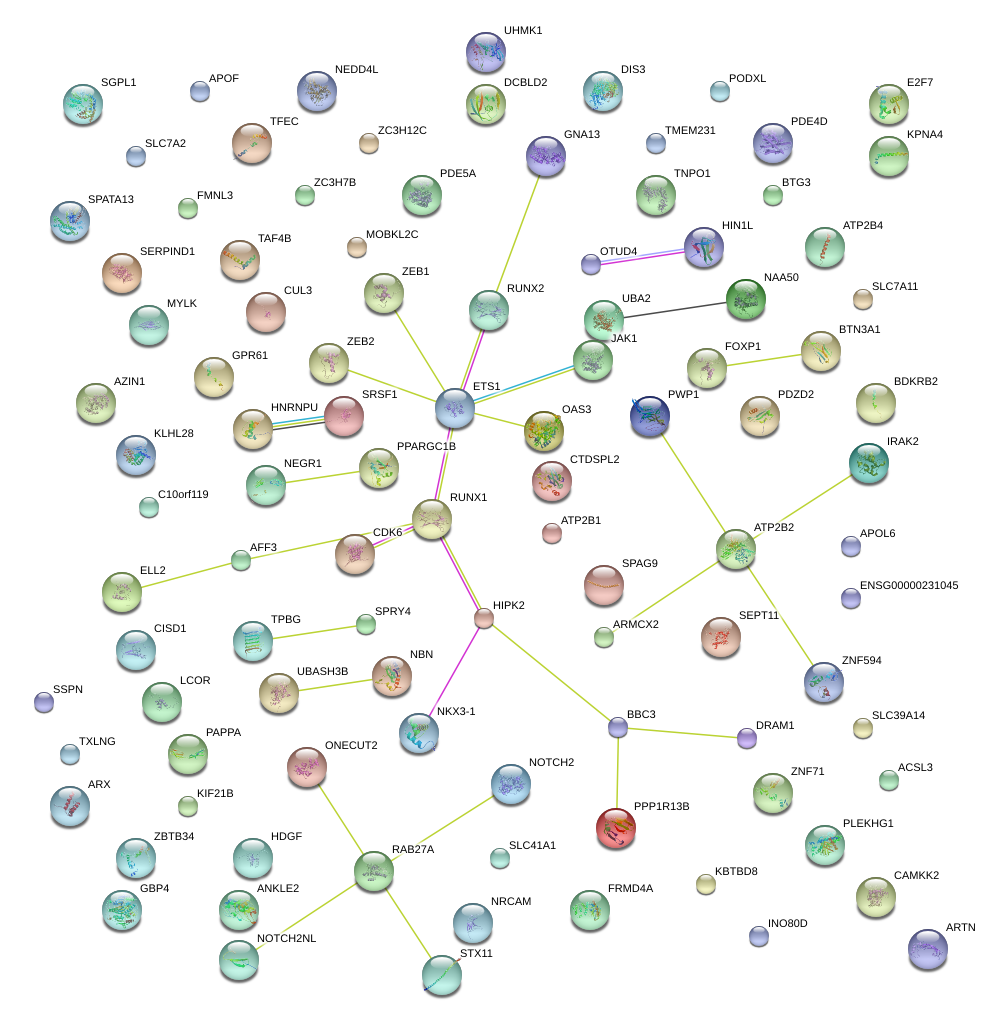
|
chr8_+_17354562
|
0.704
|
NM_001008539
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr8_+_17396285
|
0.688
|
NM_001164771
NM_003046
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr12_+_102271082
|
0.613
|
NM_018370
|
DRAM1
|
DNA-damage regulated autophagy modulator 1
|
|
chr4_-_120549115
|
0.554
|
NM_033437
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific
|
|
chr5_-_59189620
|
0.534
|
NM_001104631
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr1_+_145209067
|
0.519
|
NM_203458
|
NOTCH2NL
|
notch 2 N-terminal like
|
|
chr7_-_92465929
|
0.506
|
NM_001145306
|
CDK6
|
cyclin-dependent kinase 6
|
|
chr7_-_115670828
|
0.482
|
NM_001018058
NM_012252
|
TFEC
|
transcription factor EC
|
|
chr5_-_58882274
|
0.475
|
NM_006203
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr7_-_92463211
|
0.472
|
NM_001259
|
CDK6
|
cyclin-dependent kinase 6
|
|
chr5_-_59783885
|
0.469
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_-_58571916
|
0.463
|
NM_001197220
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_-_59064437
|
0.455
|
NM_001197218
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_-_58295758
|
0.451
|
NM_001197223
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_-_58334936
|
0.428
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_-_58652671
|
0.418
|
NM_001197219
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr4_-_120549827
|
0.401
|
NM_001083
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific
|
|
chr11_-_128457452
|
0.384
|
NM_001143820
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr16_-_75590107
|
0.372
|
NM_001077416
NM_001077418
NM_001077419
|
TMEM231
|
transmembrane protein 231
|
|
chr7_-_139477437
|
0.372
|
NM_001113239
NM_022740
|
HIPK2
|
homeodomain interacting protein kinase 2
|
|
chr11_-_128392061
|
0.365
|
NM_001162422
NM_005238
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr1_-_205782135
|
0.354
|
NM_173854
|
SLC41A1
|
solute carrier family 41, member 1
|
|
chr1_-_89664510
|
0.346
|
NM_052941
|
GBP4
|
guanylate binding protein 4
|
|
chr4_-_120548329
|
0.325
|
NM_033430
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific
|
|
chr3_-_123603144
|
0.322
|
NM_053025
NM_053026
NM_053027
NM_053028
|
MYLK
|
myosin light chain kinase
|
|
chr7_-_108096693
|
0.314
|
NM_001193582
NM_001193583
NM_001193584
NM_005010
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr3_+_10206562
|
0.313
|
NM_001570
|
IRAK2
|
interleukin-1 receptor-associated kinase 2
|
|
chr2_-_100721792
|
0.304
|
NM_001025108
|
AFF3
|
AF4/FMR2 family, member 3
|
|
chr5_-_95297597
|
0.302
|
NM_012081
|
ELL2
|
elongation factor, RNA polymerase II, 2
|
|
chr22_+_41697452
|
0.295
|
NM_017590
|
ZC3H7B
|
zinc finger CCCH-type containing 7B
|
|
chr3_-_123339423
|
0.291
|
NM_053031
NM_053032
|
MYLK
|
myosin light chain kinase
|
|
chr21_-_36421586
|
0.283
|
NM_001754
|
RUNX1
|
runt-related transcription factor 1
|
|
chr13_-_73356070
|
0.271
|
NM_001128226
NM_014953
|
DIS3
|
DIS3 mitotic control homolog (S. cerevisiae)
|
|
chr19_+_57106622
|
0.260
|
NM_021216
|
ZNF71
|
zinc finger protein 71
|
|
chr2_-_100758970
|
0.258
|
NM_002285
|
AFF3
|
AF4/FMR2 family, member 3
|
|
chr3_-_160283361
|
0.256
|
NM_002268
|
KPNA4
|
karyopherin alpha 4 (importin alpha 3)
|
|
chr14_+_96671015
|
0.254
|
NM_000623
|
BDKRB2
|
bradykinin receptor B2
|
|
chr1_+_162466963
|
0.249
|
NM_001184763
|
UHMK1
|
U2AF homology motif (UHM) kinase 1
|
|
chr10_+_60028885
|
0.249
|
NM_018464
|
CISD1
|
CDGSH iron sulfur domain 1
|
|
chrX_-_25034064
|
0.244
|
NM_139058
|
ARX
|
aristaless related homeobox
|
|
chr3_-_98620452
|
0.243
|
NM_080927
|
DCBLD2
|
discoidin, CUB and LCCL domain containing 2
|
|
chr10_+_98592658
|
0.239
|
NM_032440
|
LCOR
|
ligand dependent nuclear receptor corepressor
|
|
chr7_-_107880492
|
0.236
|
NM_001037132
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr15_-_55562483
|
0.229
|
NM_183234
NM_004580
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr11_+_122526339
|
0.228
|
NM_032873
|
UBASH3B
|
ubiquitin associated and SH3 domain containing B
|
|
chr10_+_98592015
|
0.228
|
NM_001170765
NM_001170766
|
LCOR
|
ligand dependent nuclear receptor corepressor
|
|
chr2_+_223725675
|
0.223
|
NM_004457
NM_203372
|
ACSL3
|
acyl-CoA synthetase long-chain family member 3
|
|
chr4_-_139163222
|
0.222
|
NM_014331
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11
|
|
chr12_+_113376226
|
0.219
|
NM_006187
|
OAS3
|
2'-5'-oligoadenylate synthetase 3, 100kDa
|
|
chr10_+_72575623
|
0.216
|
NM_003901
|
SGPL1
|
sphingosine-1-phosphate lyase 1
|
|
chr15_-_55563091
|
0.216
|
NM_183236
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr12_-_50101192
|
0.209
|
NM_175736
NM_198900
|
FMNL3
|
formin-like 3
|
|
chr4_+_77870864
|
0.208
|
NM_018243
|
SEPT11
|
septin 11
|
|
chr9_+_129622935
|
0.207
|
NM_001099270
|
ZBTB34
|
zinc finger and BTB domain containing 34
|
|
chr6_+_26402464
|
0.206
|
NM_001145008
NM_001145009
NM_007048
NM_194441
|
BTN3A1
|
butyrophilin, subfamily 3, member A1
|
|
chr11_+_109964086
|
0.202
|
NM_033390
|
ZC3H12C
|
zinc finger CCCH-type containing 12C
|
|
chr1_+_162467594
|
0.202
|
NM_144624
NM_175866
|
UHMK1
|
U2AF homology motif (UHM) kinase 1
|
|
chr8_-_90996884
|
0.201
|
NM_002485
|
NBN
|
nibrin
|
|
chr6_+_144471631
|
0.201
|
NM_003764
|
STX11
|
syntaxin 11
|
|
chr19_-_47734215
|
0.200
|
NM_014417
|
BBC3
|
BCL2 binding component 3
|
|
chr10_-_14372807
|
0.200
|
NM_018027
|
FRMD4A
|
FERM domain containing 4A
|
|
chr12_-_121734543
|
0.198
|
NM_172226
|
CAMKK2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta
|
|
chr15_-_55581969
|
0.197
|
NM_183235
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr1_-_156722023
|
0.196
|
NM_001126051
NM_001126050
|
HDGF
|
hepatoma-derived growth factor
|
|
chr8_-_23540401
|
0.195
|
NM_006167
|
NKX3-1
|
NK3 homeobox 1
|
|
chr1_-_72748147
|
0.195
|
NM_173808
|
NEGR1
|
neuronal growth regulator 1
|
|
chr18_+_23806385
|
0.193
|
NM_005640
|
TAF4B
|
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa
|
|
chr3_-_113465101
|
0.192
|
NM_025146
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit
|
|
chr1_-_156721501
|
0.191
|
NM_004494
|
HDGF
|
hepatoma-derived growth factor
|
|
chr21_-_18985224
|
0.189
|
NM_001130914
NM_006806
|
BTG3
|
BTG family, member 3
|
|
chr22_+_36044412
|
0.188
|
NM_030641
|
APOL6
|
apolipoprotein L, 6
|
|
chr6_+_83073954
|
0.184
|
NM_001166392
|
TPBG
|
trophoblast glycoprotein
|
|
chr3_-_10547267
|
0.183
|
NM_001001331
NM_001683
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr12_+_26348268
|
0.183
|
NM_001135823
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr6_+_83072922
|
0.179
|
NM_006670
|
TPBG
|
trophoblast glycoprotein
|
|
chr6_-_13295817
|
0.172
|
NM_001242698
|
LOC100130357
|
uncharacterized LOC100130357
|
|
chr14_-_45431121
|
0.171
|
NM_017658
|
KLHL28
|
kelch-like 28 (Drosophila)
|
|
chr13_+_24734788
|
0.169
|
NM_001166271
NM_153023
|
SPATA13
|
spermatogenesis associated 13
|
|
chr8_+_22224997
|
0.168
|
NM_001135153
|
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14
|
|
chr1_+_110082493
|
0.163
|
NM_031936
|
GPR61
|
G protein-coupled receptor 61
|
|
chr6_+_150920998
|
0.163
|
NM_001029884
|
PLEKHG1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1
|
|
chr8_-_103876390
|
0.163
|
NM_015878
NM_148174
|
AZIN1
|
antizyme inhibitor 1
|
|
chr22_+_21128378
|
0.162
|
NM_000185
|
SERPIND1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1
|
|
chr1_-_65431826
|
0.160
|
NM_002227
|
JAK1
|
Janus kinase 1
|
|
chr2_-_145277879
|
0.159
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr1_-_47082529
|
0.158
|
NM_201403
|
MOB3C
|
MOB kinase activator 3C
|
|
chr12_-_77459198
|
0.157
|
NM_203394
|
E2F7
|
E2F transcription factor 7
|
|
chr1_-_47080793
|
0.157
|
NM_145279
|
MOB3C
|
MOB kinase activator 3C
|
|
chr5_+_149109814
|
0.157
|
NM_001172698
NM_133263
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr5_+_31799030
|
0.157
|
NM_178140
|
PDZD2
|
PDZ domain containing 2
|
|
chrX_+_16804531
|
0.156
|
NM_001168683
NM_018360
|
TXLNG
|
taxilin gamma
|
|
chr3_+_119187782
|
0.156
|
NM_152305
|
POGLUT1
|
protein O-glucosyltransferase 1
|
|
chr3_+_67048726
|
0.155
|
NM_032505
|
KBTBD8
|
kelch repeat and BTB (POZ) domain containing 8
|
|
chr7_-_131241321
|
0.153
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr3_-_71632741
|
0.152
|
NM_001244808
NM_001012505
NM_001244810
NM_032682
|
FOXP1
|
forkhead box P1
|
|
chr5_-_141704575
|
0.151
|
NM_001127496
NM_030964
|
SPRY4
|
sprouty homolog 4 (Drosophila)
|
|
chr15_+_44719578
|
0.151
|
NM_016396
|
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2
|
|
chr17_-_56084656
|
0.150
|
NM_001078166
NM_006924
|
SRSF1
|
serine/arginine-rich splicing factor 1
|
|
chr18_+_55862577
|
0.149
|
NM_001144970
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr2_-_206950895
|
0.148
|
NM_017759
|
INO80D
|
INO80 complex subunit D
|
|
chr17_-_49198084
|
0.148
|
NM_001130527
NM_001130528
NM_003971
|
SPAG9
|
sperm associated antigen 9
|
|
chr10_-_121632265
|
0.147
|
NM_024834
|
MCMBP
|
minichromosome maintenance complex binding protein
|
|
chr14_-_104313730
|
0.145
|
NM_015316
|
PPP1R13B
|
protein phosphatase 1, regulatory subunit 13B
|
|
chr18_+_55711388
|
0.145
|
NM_001144967
NM_001243960
NM_015277
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr17_-_63052884
|
0.144
|
NM_006572
|
GNA13
|
guanine nucleotide binding protein (G protein), alpha 13
|
|
chr1_-_245027808
|
0.144
|
NM_004501
NM_031844
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A)
|
|
chr4_-_146100793
|
0.143
|
NM_001102653
|
OTUD4
|
OTU domain containing 4
|
|
chr2_-_225450105
|
0.143
|
NM_003590
|
CUL3
|
cullin 3
|
|
chr12_-_133338389
|
0.142
|
NM_015114
|
ANKLE2
|
ankyrin repeat and LEM domain containing 2
|
|
chr5_+_72112417
|
0.141
|
NM_002270
|
TNPO1
|
transportin 1
|
|
chr18_+_55714457
|
0.139
|
NM_001144964
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr18_+_55888750
|
0.136
|
NM_001144966
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr18_+_55102777
|
0.135
|
NM_004852
|
ONECUT2
|
one cut homeobox 2
|
|
chr1_-_200992824
|
0.135
|
NM_001252100
NM_001252102
NM_001252103
NM_017596
|
KIF21B
|
kinesin family member 21B
|
|
chr1_+_25071759
|
0.135
|
NM_013943
|
CLIC4
|
chloride intracellular channel 4
|
|
chr12_+_26348488
|
0.133
|
NM_005086
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr3_-_69101236
|
0.132
|
NM_007114
|
TMF1
|
TATA element modulatory factor 1
|
|
chr8_-_105601234
|
0.131
|
NM_001135703
NM_013437
|
LRP12
|
low density lipoprotein receptor-related protein 12
|
|
chr1_-_55681024
|
0.130
|
NM_015306
|
USP24
|
ubiquitin specific peptidase 24
|
|
chr19_-_17958814
|
0.130
|
NM_000215
|
JAK3
|
Janus kinase 3
|
|
chr9_+_80911990
|
0.130
|
NM_021154
NM_058179
|
PSAT1
|
phosphoserine aminotransferase 1
|
|
chr15_+_100106132
|
0.128
|
NM_001130926
NM_001130927
NM_001171894
NM_005587
|
MEF2A
|
myocyte enhancer factor 2A
|
|
chr5_+_179233351
|
0.127
|
NM_001142298
|
SQSTM1
|
sequestosome 1
|
|
chr10_+_60094712
|
0.126
|
NM_001204880
NM_003338
|
UBE2D1
|
ubiquitin-conjugating enzyme E2D 1
|
|
chr2_-_153573858
|
0.125
|
NM_017892
|
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae)
|
|
chr5_+_72143929
|
0.123
|
NM_153188
|
TNPO1
|
transportin 1
|
|
chr18_+_55816546
|
0.123
|
NM_001144965
NM_001144968
NM_001144969
NM_001144971
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr6_+_37787266
|
0.123
|
NM_021943
|
ZFAND3
|
zinc finger, AN1-type domain 3
|
|
chr7_+_129932973
|
0.123
|
NM_001163446
NM_016352
|
CPA4
|
carboxypeptidase A4
|
|
chr2_-_11484699
|
0.121
|
NM_004850
|
ROCK2
|
Rho-associated, coiled-coil containing protein kinase 2
|
|
chr17_+_27717942
|
0.121
|
NM_020791
NM_025142
|
TAOK1
|
TAO kinase 1
|
|
chr15_+_85525204
|
0.119
|
NM_002605
NM_173454
|
PDE8A
|
phosphodiesterase 8A
|
|
chr3_+_186648273
|
0.118
|
NM_173216
NM_173217
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chrX_-_40506800
|
0.118
|
NM_144970
|
CXorf38
|
chromosome X open reading frame 38
|
|
chr15_+_100173182
|
0.117
|
NM_001130928
|
MEF2A
|
myocyte enhancer factor 2A
|
|
chr16_+_31191430
|
0.117
|
NM_001170634
NM_001170937
NM_004960
|
FUS
|
fused in sarcoma
|
|
chr19_+_51815095
|
0.116
|
NM_001101372
|
IGLON5
|
IgLON family member 5
|
|
chr17_-_28257017
|
0.113
|
NM_033389
|
SSH2
|
slingshot homolog 2 (Drosophila)
|
|
chr5_+_179234002
|
0.113
|
NM_001142299
|
SQSTM1
|
sequestosome 1
|
|
chr16_-_72128214
|
0.113
|
NM_001142318
|
TXNL4B
|
thioredoxin-like 4B
|
|
chr16_-_72127455
|
0.112
|
NM_017853
|
TXNL4B
|
thioredoxin-like 4B
|
|
chr3_+_186739643
|
0.111
|
NM_003032
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr16_+_25703346
|
0.111
|
NM_006040
|
HS3ST4
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 4
|
|
chr11_+_32914359
|
0.111
|
NM_001076786
|
QSER1
|
glutamine and serine rich 1
|
|
chr5_+_148651395
|
0.110
|
NM_001146337
NM_152406
|
AFAP1L1
|
actin filament associated protein 1-like 1
|
|
chr1_+_2985730
|
0.109
|
NM_022114
NM_199454
|
PRDM16
|
PR domain containing 16
|
|
chr8_+_81398416
|
0.108
|
NM_001105539
NM_023929
|
ZBTB10
|
zinc finger and BTB domain containing 10
|
|
chr16_-_72127767
|
0.107
|
NM_001142317
|
TXNL4B
|
thioredoxin-like 4B
|
|
chr17_-_42907606
|
0.106
|
NM_005497
|
GJC1
|
gap junction protein, gamma 1, 45kDa
|
|
chr1_+_167691156
|
0.106
|
NM_001146191
NM_003953
NM_024569
|
MPZL1
|
myelin protein zero-like 1
|
|
chr2_+_192542860
|
0.106
|
NM_001031716
|
OBFC2A
|
oligonucleotide/oligosaccharide-binding fold containing 2A
|
|
chr9_+_102668904
|
0.105
|
NM_017919
|
STX17
|
syntaxin 17
|
|
chr7_-_141401889
|
0.105
|
NM_001080392
|
KIAA1147
|
KIAA1147
|
|
chr9_+_135545727
|
0.104
|
NM_012204
|
GTF3C4
|
general transcription factor IIIC, polypeptide 4, 90kDa
|
|
chr20_+_62526453
|
0.104
|
NM_025219
|
DNAJC5
|
DnaJ (Hsp40) homolog, subfamily C, member 5
|
|
chr5_+_179247841
|
0.101
|
NM_003900
|
SQSTM1
|
sequestosome 1
|
|
chr5_+_170846659
|
0.101
|
NM_003862
|
FGF18
|
fibroblast growth factor 18
|
|
chr12_+_12764755
|
0.101
|
NM_001310
|
CREBL2
|
cAMP responsive element binding protein-like 2
|
|
chr10_-_43892697
|
0.101
|
NM_001098207
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F
|
|
chr18_+_49866541
|
0.100
|
NM_005215
|
DCC
|
deleted in colorectal carcinoma
|
|
chr1_+_167298280
|
0.099
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr6_-_89927495
|
0.098
|
NM_002042
|
GABRR1
|
gamma-aminobutyric acid (GABA) receptor, rho 1
|
|
chr5_-_114880536
|
0.098
|
NM_020177
|
FEM1C
|
fem-1 homolog c (C. elegans)
|
|
chr7_+_107384278
|
0.097
|
NM_024814
|
CBLL1
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence-like 1
|
|
chr10_-_43903220
|
0.097
|
NM_001098206
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F
|
|
chr8_-_124054591
|
0.096
|
NM_001134671
NM_024295
|
DERL1
|
Der1-like domain family, member 1
|
|
chr3_+_160939098
|
0.096
|
NM_015938
|
NMD3
|
NMD3 homolog (S. cerevisiae)
|
|
chr10_+_134000335
|
0.096
|
NM_006426
|
DPYSL4
|
dihydropyrimidinase-like 4
|
|
chr5_-_142783975
|
0.095
|
NM_001018076
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr9_-_117568292
|
0.095
|
NM_005118
|
TNFSF15
|
tumor necrosis factor (ligand) superfamily, member 15
|
|
chr19_-_47735858
|
0.095
|
NM_001127240
NM_001127241
NM_001127242
|
BBC3
|
BCL2 binding component 3
|
|
chr10_+_27793161
|
0.095
|
NM_021252
|
RAB18
|
RAB18, member RAS oncogene family
|
|
chr6_+_34759743
|
0.095
|
NM_017754
|
UHRF1BP1
|
UHRF1 binding protein 1
|
|
chr2_+_54785453
|
0.094
|
NM_178313
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1
|
|
chr7_+_20370724
|
0.094
|
NM_002214
|
ITGB8
|
integrin, beta 8
|
|
chr13_+_97874540
|
0.094
|
NM_144778
NM_207304
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr17_-_42908178
|
0.093
|
NM_001080383
|
GJC1
|
gap junction protein, gamma 1, 45kDa
|
|
chrX_-_142722869
|
0.093
|
NM_173078
|
SLITRK4
|
SLIT and NTRK-like family, member 4
|
|
chr1_+_114493766
|
0.093
|
NM_198269
|
HIPK1
|
homeodomain interacting protein kinase 1
|
|
chr16_-_12897693
|
0.093
|
NM_001099455
NM_018340
|
CPPED1
|
calcineurin-like phosphoesterase domain containing 1
|
|
chr2_+_166428845
|
0.092
|
NM_024969
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr13_+_25946084
|
0.092
|
NM_016529
|
ATP8A2
|
ATPase, aminophospholipid transporter, class I, type 8A, member 2
|
|
chr2_+_73144603
|
0.092
|
NM_004097
|
EMX1
|
empty spiracles homeobox 1
|
|
chr5_-_132073143
|
0.092
|
NM_007054
|
KIF3A
|
kinesin family member 3A
|
|
chr16_-_755718
|
0.091
|
NM_153350
|
FBXL16
|
F-box and leucine-rich repeat protein 16
|
|
chr10_+_73723992
|
0.090
|
NM_004273
|
CHST3
|
carbohydrate (chondroitin 6) sulfotransferase 3
|
|
chr22_-_36236421
|
0.090
|
NM_001031695
NM_001082576
NM_001082577
NM_014309
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr15_+_99192696
|
0.090
|
NM_000875
|
IGF1R
|
insulin-like growth factor 1 receptor
|
|
chr19_-_54604070
|
0.090
|
NM_130771
NM_133168
NM_133169
NM_206818
|
OSCAR
|
osteoclast associated, immunoglobulin-like receptor
|
|
chr3_+_29322802
|
0.089
|
NM_001003792
NM_001003793
NM_001177711
NM_001177712
NM_014483
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3
|
|
chr12_+_70132562
|
0.089
|
NM_022456
NM_175624
|
RAB3IP
|
RAB3A interacting protein (rabin3)
|
|
chr2_-_202316227
|
0.088
|
NM_015049
|
TRAK2
|
trafficking protein, kinesin binding 2
|
|
chr10_-_43892160
|
0.088
|
NM_001098208
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F
|
|
chr5_+_56110899
|
0.088
|
NM_005921
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1
|
|
chr14_+_75894508
|
0.087
|
NM_001135047
|
JDP2
|
Jun dimerization protein 2
|
|
chr16_+_2587951
|
0.087
|
NM_002613
NM_031268
|
PDPK1
|
3-phosphoinositide dependent protein kinase-1
|
|
chr2_+_27070962
|
0.087
|
NM_020134
NM_001253723
NM_001253724
|
DPYSL5
|
dihydropyrimidinase-like 5
|
|
chr10_-_43904306
|
0.087
|
NM_001098205
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F
|
|
chr3_-_119812973
|
0.086
|
NM_001146156
NM_002093
|
GSK3B
|
glycogen synthase kinase 3 beta
|
|
chr5_-_16936371
|
0.086
|
NM_012334
|
MYO10
|
myosin X
|