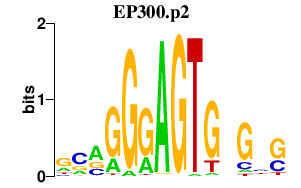
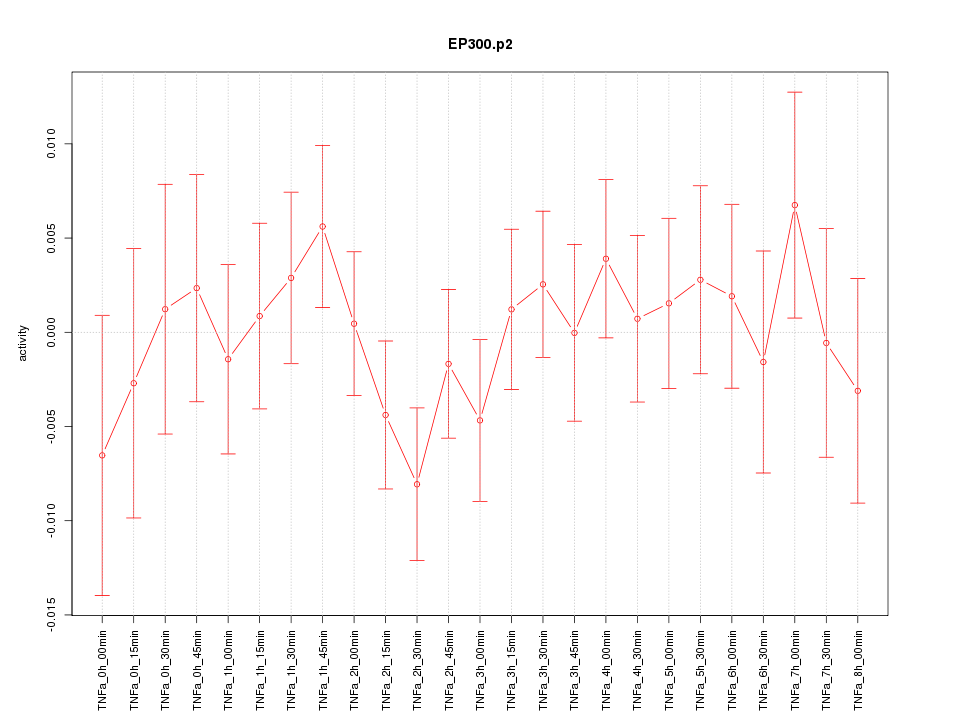
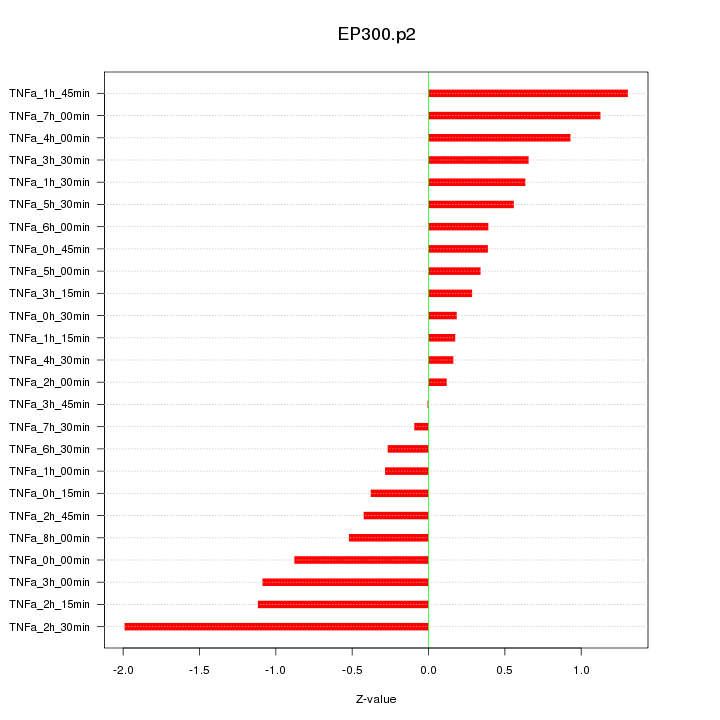
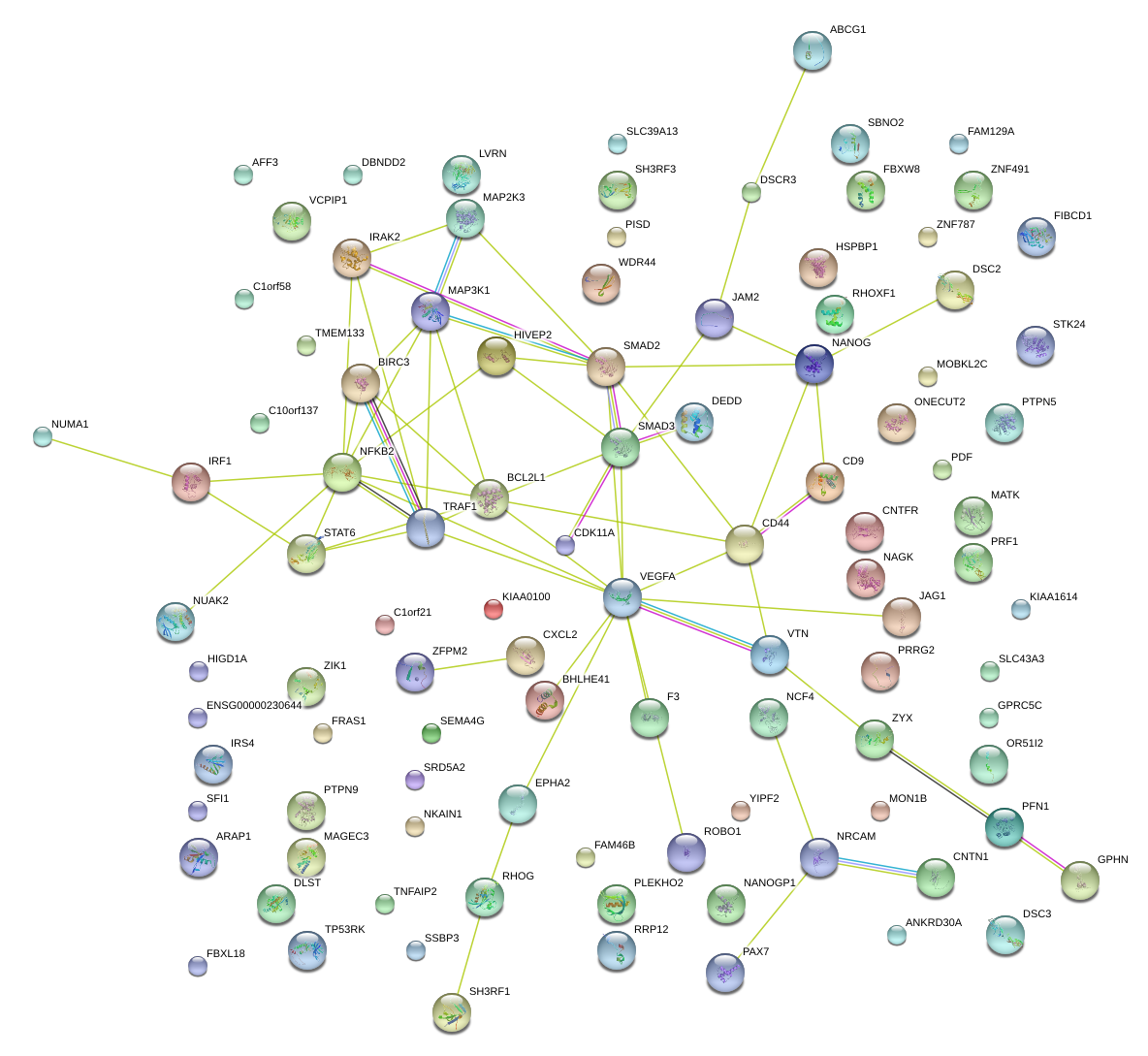
|
chr1_-_95007139
|
1.489
|
NM_001178096
NM_001993
|
F3
|
coagulation factor III (thromboplastin, tissue factor)
|
|
chr5_-_131826426
|
0.814
|
NM_002198
|
IRF1
|
interferon regulatory factor 1
|
|
chr9_-_123688352
|
0.725
|
|
TRAF1
|
TNF receptor-associated factor 1
|
|
chr16_+_77224865
|
0.582
|
|
MON1B
|
MON1 homolog B (yeast)
|
|
chr1_-_184943673
|
0.544
|
NM_052966
|
FAM129A
|
family with sequence similarity 129, member A
|
|
chr11_+_102188180
|
0.534
|
NM_001165
NM_182962
|
BIRC3
|
baculoviral IAP repeat containing 3
|
|
chr21_+_46293925
|
0.444
|
|
|
|
|
chr8_+_106331146
|
0.433
|
NM_012082
|
ZFPM2
|
zinc finger protein, multitype 2
|
|
chr16_+_77224827
|
0.424
|
NM_014940
|
MON1B
|
MON1 homolog B (yeast)
|
|
chr12_-_26277807
|
0.412
|
NM_030762
|
BHLHE41
|
basic helix-loop-helix family, member e41
|
|
chr16_+_77225107
|
0.407
|
|
MON1B
|
MON1 homolog B (yeast)
|
|
chr3_-_79817058
|
0.375
|
NM_002941
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr19_-_56632644
|
0.354
|
NM_001002836
|
ZNF787
|
zinc finger protein 787
|
|
chr5_+_56111379
|
0.337
|
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1
|
|
chr17_-_4851688
|
0.334
|
|
PFN1
|
profilin 1
|
|
chr2_-_100721792
|
0.326
|
NM_001025108
|
AFF3
|
AF4/FMR2 family, member 3
|
|
chr4_-_38666429
|
0.321
|
|
FLJ13197
|
uncharacterized FLJ13197
|
|
chrX_+_140982451
|
0.316
|
NM_177456
|
MAGEC3
|
melanoma antigen family C, 3
|
|
chr3_+_10206562
|
0.316
|
NM_001570
|
IRAK2
|
interleukin-1 receptor-associated kinase 2
|
|
chr11_-_57194556
|
0.313
|
|
SLC43A3
|
solute carrier family 43, member 3
|
|
chr1_+_222886728
|
0.312
|
|
BROX
|
BRO1 domain and CAAX motif containing
|
|
chr1_+_180882312
|
0.310
|
NM_020950
|
KIAA1614
|
KIAA1614
|
|
chr17_-_4851797
|
0.310
|
NM_005022
|
PFN1
|
profilin 1
|
|
chr12_+_7942025
|
0.310
|
|
NANOG
|
Nanog homeobox
|
|
chr1_+_18957499
|
0.308
|
NM_001135254
NM_002584
NM_013945
|
PAX7
|
paired box 7
|
|
chr20_-_10654429
|
0.305
|
NM_000214
|
JAG1
|
jagged 1
|
|
chr19_-_11039086
|
0.303
|
NM_024029
|
YIPF2
|
Yip1 domain family, member 2
|
|
chr1_-_161102367
|
0.297
|
|
DEDD
|
death effector domain containing
|
|
chr17_-_26697310
|
0.295
|
|
VTN
|
vitronectin
|
|
chr11_-_3862137
|
0.294
|
|
RHOG
|
ras homolog gene family, member G (rho G)
|
|
chr4_-_121993568
|
0.289
|
NM_024574
|
NDNF
|
neuron-derived neurotrophic factor
|
|
chr1_-_184943532
|
0.284
|
|
FAM129A
|
family with sequence similarity 129, member A
|
|
chr11_-_71791734
|
0.279
|
|
NUMA1
|
nuclear mitotic apparatus protein 1
|
|
chr12_+_117348710
|
0.277
|
NM_012174
NM_153348
|
FBXW8
|
F-box and WD repeat domain containing 8
|
|
chr1_-_161102457
|
0.277
|
NM_001039711
NM_032998
|
DEDD
|
death effector domain containing
|
|
chr17_-_4851827
|
0.274
|
|
PFN1
|
profilin 1
|
|
chr11_+_5474637
|
0.273
|
NM_001004754
|
OR51I2
|
olfactory receptor, family 51, subfamily I, member 2
|
|
chr14_+_66974848
|
0.273
|
|
GPHN
|
gephyrin
|
|
chr4_-_74964996
|
0.271
|
NM_002089
|
CXCL2
|
chemokine (C-X-C motif) ligand 2
|
|
chr14_+_75348592
|
0.266
|
NM_001244883
NM_001933
|
DLST
|
dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex)
|
|
chr13_-_99229037
|
0.264
|
|
STK24
|
serine/threonine kinase 24
|
|
chr1_-_1590403
|
0.263
|
|
CDK11A
CDK11B
|
cyclin-dependent kinase 11A
cyclin-dependent kinase 11B
|
|
chr10_+_102729249
|
0.263
|
|
SEMA4G
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G
|
|
chr9_-_34590137
|
0.260
|
NM_001207011
|
CNTFR
|
ciliary neurotrophic factor receptor
|
|
chr20_-_30311693
|
0.258
|
|
BCL2L1
|
BCL2-like 1
|
|
chr1_-_161102210
|
0.257
|
NM_001039712
|
DEDD
|
death effector domain containing
|
|
chr11_-_57194591
|
0.256
|
NM_199329
|
SLC43A3
|
solute carrier family 43, member 3
|
|
chr19_-_1174263
|
0.245
|
NM_014963
|
SBNO2
|
strawberry notch homolog 2 (Drosophila)
|
|
chr15_+_67358194
|
0.244
|
NM_005902
|
SMAD3
|
SMAD family member 3
|
|
chr10_+_37414784
|
0.244
|
NM_052997
|
ANKRD30A
|
ankyrin repeat domain 30A
|
|
chr1_-_16482426
|
0.243
|
NM_004431
|
EPHA2
|
EPH receptor A2
|
|
chr7_-_5553401
|
0.241
|
|
FBXL18
|
F-box and leucine-rich repeat protein 18
|
|
chr6_-_143266283
|
0.240
|
NM_006734
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2
|
|
chr7_+_143078395
|
0.239
|
|
ZYX
|
zyxin
|
|
chr6_+_43738757
|
0.239
|
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr21_-_38639593
|
0.236
|
|
DSCR3
|
Down syndrome critical region gene 3
|
|
chr11_-_57194512
|
0.236
|
NM_014096
|
SLC43A3
|
solute carrier family 43, member 3
|
|
chr20_-_45318135
|
0.231
|
NM_033550
|
TP53RK
|
TP53 regulating kinase
|
|
chr17_-_26697224
|
0.230
|
|
VTN
|
vitronectin
|
|
chr7_+_143078358
|
0.230
|
NM_001010972
NM_003461
|
ZYX
|
zyxin
|
|
chr11_+_35160385
|
0.230
|
NM_000610
NM_001001389
NM_001001390
NM_001001391
NM_001001392
NM_001202555
NM_001202556
NM_001202557
|
CD44
|
CD44 molecule (Indian blood group)
|
|
chr1_+_222886655
|
0.222
|
|
BROX
|
BRO1 domain and CAAX motif containing
|
|
chr12_+_6309475
|
0.220
|
NM_001769
|
CD9
|
CD9 molecule
|
|
chr18_+_55102777
|
0.218
|
NM_004852
|
ONECUT2
|
one cut homeobox 2
|
|
chr2_+_71295685
|
0.215
|
|
NAGK
|
N-acetylglucosamine kinase
|
|
chr8_-_67579362
|
0.214
|
|
VCPIP1
|
valosin containing protein (p97)/p47 complex interacting protein 1
|
|
chr17_+_21188021
|
0.212
|
|
MAP2K3
|
mitogen-activated protein kinase kinase 3
|
|
chrX_+_117480416
|
0.212
|
|
WDR44
|
WD repeat domain 44
|
|
chr14_+_103592663
|
0.212
|
NM_006291
|
TNFAIP2
|
tumor necrosis factor, alpha-induced protein 2
|
|
chr10_+_127408225
|
0.211
|
|
C10orf137
|
chromosome 10 open reading frame 137
|
|
chr22_+_37257004
|
0.211
|
NM_000631
NM_013416
|
NCF4
|
neutrophil cytosolic factor 4, 40kDa
|
|
chr1_-_205290756
|
0.210
|
NM_030952
|
NUAK2
|
NUAK family, SNF1-like kinase, 2
|
|
chr17_-_26972165
|
0.209
|
|
KIAA0100
|
KIAA0100
|
|
chr8_-_67579377
|
0.207
|
|
VCPIP1
|
valosin containing protein (p97)/p47 complex interacting protein 1
|
|
chr20_+_44035203
|
0.206
|
NM_001048223
NM_001048224
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2
|
|
chr1_-_47082529
|
0.206
|
NM_201403
|
MOB3C
|
MOB kinase activator 3C
|
|
chr19_+_11909360
|
0.200
|
NM_152356
|
ZNF491
|
zinc finger protein 491
|
|
chr9_-_133814236
|
0.198
|
NM_032843
|
FIBCD1
|
fibrinogen C domain containing 1
|
|
chr17_+_72427535
|
0.196
|
NM_022036
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C
|
|
chr11_-_18813127
|
0.195
|
NM_001039970
NM_006906
NM_032781
|
PTPN5
|
protein tyrosine phosphatase, non-receptor type 5 (striatum-enriched)
|
|
chr20_+_44034632
|
0.193
|
NM_001197139
NM_001048221
NM_001048222
NM_001197140
NM_018478
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2
|
|
chr15_-_75871626
|
0.191
|
|
PTPN9
|
protein tyrosine phosphatase, non-receptor type 9
|
|
chr15_+_65134081
|
0.190
|
NM_001195059
NM_025201
|
PLEKHO2
|
pleckstrin homology domain containing, family O member 2
|
|
chr17_-_26697293
|
0.189
|
|
VTN
|
vitronectin
|
|
chr2_-_31805968
|
0.189
|
NM_000348
|
SRD5A2
|
steroid-5-alpha-reductase, alpha polypeptide 2 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 2)
|
|
chr1_-_54871681
|
0.188
|
|
SSBP3
|
single stranded DNA binding protein 3
|
|
chr10_+_104155431
|
0.187
|
NM_002502
|
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100)
|
|
chr7_+_143078457
|
0.187
|
|
ZYX
|
zyxin
|
|
chr3_-_79068542
|
0.186
|
NM_001145845
NM_133631
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr22_-_32026783
|
0.186
|
NM_014338
|
PISD
|
phosphatidylserine decarboxylase
|
|
chr2_+_109745996
|
0.186
|
NM_001099289
|
SH3RF3
|
SH3 domain containing ring finger 3
|
|
chr19_+_50084586
|
0.185
|
NM_000951
|
PRRG2
|
proline rich Gla (G-carboxyglutamic acid) 2
|
|
chr1_+_184356186
|
0.184
|
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr7_+_143078399
|
0.184
|
|
ZYX
|
zyxin
|
|
chr21_+_43639202
|
0.184
|
NM_004915
NM_016818
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1
|
|
chr1_-_31712733
|
0.183
|
NM_024522
|
NKAIN1
|
Na+/K+ transporting ATPase interacting 1
|
|
chrX_-_119249774
|
0.183
|
NM_139282
|
RHOXF1
|
Rhox homeobox family, member 1
|
|
chr19_-_55791727
|
0.183
|
NM_001130106
NM_012267
|
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1
|
|
chr19_-_3786221
|
0.182
|
|
MATK
|
megakaryocyte-associated tyrosine kinase
|
|
chr1_-_27339448
|
0.182
|
|
FAM46B
|
family with sequence similarity 46, member B
|
|
chr11_-_72433379
|
0.182
|
NM_001135190
NM_015242
|
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1
|
|
chr19_-_47104456
|
0.182
|
NM_001205281
|
LOC100506012
|
serine/threonine-protein phosphatase 5-like
|
|
chr20_+_9494995
|
0.181
|
NM_001199897
NM_012261
|
LAMP5
|
lysosomal-associated membrane protein family, member 5
|
|
chr21_+_27011588
|
0.180
|
NM_021219
|
JAM2
|
junctional adhesion molecule 2
|
|
chr4_+_78978723
|
0.179
|
NM_001166133
NM_025074
|
FRAS1
|
Fraser syndrome 1
|
|
chr16_-_69373473
|
0.178
|
NM_032382
|
COG8
|
component of oligomeric golgi complex 8
|
|
chr10_-_99161013
|
0.177
|
NM_001145114
NM_015179
|
RRP12
|
ribosomal RNA processing 12 homolog (S. cerevisiae)
|
|
chr11_+_47430045
|
0.177
|
NM_001128225
NM_152264
|
SLC39A13
|
solute carrier family 39 (zinc transporter), member 13
|
|
chr18_-_28681885
|
0.176
|
NM_004949
NM_024422
|
DSC2
|
desmocollin 2
|
|
chr12_-_57505121
|
0.175
|
|
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced
|
|
chr7_-_108096693
|
0.175
|
NM_001193582
NM_001193583
NM_001193584
NM_005010
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chrX_-_107979606
|
0.174
|
NM_003604
|
IRS4
|
insulin receptor substrate 4
|
|
chr5_+_115298136
|
0.173
|
NM_173800
|
AQPEP
|
laeverin
|
|
chr22_-_17602142
|
0.173
|
NM_031890
NM_001163079
|
CECR6
|
cat eye syndrome chromosome region, candidate 6
|
|
chr19_-_55791446
|
0.172
|
|
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1
|
|
chr15_-_75871508
|
0.172
|
NM_002833
|
PTPN9
|
protein tyrosine phosphatase, non-receptor type 9
|
|
chr11_-_65667809
|
0.171
|
|
FOSL1
|
FOS-like antigen 1
|
|
chr2_-_225907312
|
0.171
|
NM_014689
|
DOCK10
|
dedicator of cytokinesis 10
|
|
chr6_-_154831704
|
0.171
|
NM_173515
|
CNKSR3
|
CNKSR family member 3
|
|
chr7_+_143078444
|
0.170
|
|
ZYX
|
zyxin
|
|
chr19_-_55791428
|
0.169
|
|
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1
|
|
chr1_+_1243971
|
0.168
|
NM_153339
|
PUSL1
|
pseudouridylate synthase-like 1
|
|
chr2_-_136743076
|
0.166
|
|
DARS
|
aspartyl-tRNA synthetase
|
|
chr11_-_65667860
|
0.165
|
NM_005438
|
FOSL1
|
FOS-like antigen 1
|
|
chr19_+_13105970
|
0.165
|
NM_002501
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr21_+_43639930
|
0.163
|
NM_207174
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1
|
|
chr20_-_10654139
|
0.163
|
|
JAG1
|
jagged 1
|
|
chr9_-_133813988
|
0.162
|
|
FIBCD1
|
fibrinogen C domain containing 1
|
|
chr19_+_39390258
|
0.162
|
NM_001243116
|
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta
|
|
chr15_+_74908195
|
0.162
|
|
CLK3
|
CDC-like kinase 3
|
|
chr2_-_136743189
|
0.161
|
NM_001349
|
DARS
|
aspartyl-tRNA synthetase
|
|
chr19_-_1174201
|
0.161
|
|
|
|
|
chr12_-_6740814
|
0.160
|
NM_001142961
|
LPAR5
|
lysophosphatidic acid receptor 5
|
|
chr6_-_31324975
|
0.160
|
NM_005514
|
HLA-B
|
major histocompatibility complex, class I, B
|
|
chr7_-_37488408
|
0.160
|
|
ELMO1
|
engulfment and cell motility 1
|
|
chr9_-_33473846
|
0.160
|
|
NOL6
|
nucleolar protein family 6 (RNA-associated)
|
|
chr5_-_150460599
|
0.160
|
NM_001252385
NM_001252386
NM_001252393
NM_006058
|
TNIP1
|
TNFAIP3 interacting protein 1
|
|
chr10_+_23983674
|
0.158
|
NM_001098500
|
KIAA1217
|
KIAA1217
|
|
chr7_+_108210308
|
0.158
|
|
DNAJB9
|
DnaJ (Hsp40) homolog, subfamily B, member 9
|
|
chr2_+_168043792
|
0.157
|
NM_001199144
NM_001199145
|
XIRP2
|
xin actin-binding repeat containing 2
|
|
chr19_-_55791490
|
0.157
|
|
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1
|
|
chr1_-_228604582
|
0.157
|
NM_001024940
NM_001134855
NM_016102
|
TRIM17
|
tripartite motif containing 17
|
|
chr4_-_185747051
|
0.157
|
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1
|
|
chr17_+_46985720
|
0.155
|
NM_023079
|
UBE2Z
|
ubiquitin-conjugating enzyme E2Z
|
|
chr18_-_28681429
|
0.154
|
|
DSC2
|
desmocollin 2
|
|
chr12_-_57505176
|
0.153
|
NM_001178078
NM_001178080
NM_001178081
NM_003153
|
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced
|
|
chr11_+_65479685
|
0.153
|
|
KAT5
|
K(lysine) acetyltransferase 5
|
|
chr1_-_228604305
|
0.152
|
|
TRIM17
|
tripartite motif containing 17
|
|
chr1_+_229761988
|
0.152
|
|
URB2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae)
|
|
chr19_+_10982369
|
0.152
|
|
CARM1
|
coactivator-associated arginine methyltransferase 1
|
|
chr3_-_158450271
|
0.152
|
NM_002888
NM_206963
|
RARRES1
|
retinoic acid receptor responder (tazarotene induced) 1
|
|
chr6_+_99282597
|
0.151
|
|
POU3F2
|
POU class 3 homeobox 2
|
|
chr5_-_150460550
|
0.150
|
|
TNIP1
|
TNFAIP3 interacting protein 1
|
|
chr17_+_46985829
|
0.150
|
|
UBE2Z
|
ubiquitin-conjugating enzyme E2Z
|
|
chr15_+_69706619
|
0.150
|
NM_004856
NM_138555
|
KIF23
|
kinesin family member 23
|
|
chr15_+_74287148
|
0.149
|
|
PML
|
promyelocytic leukemia
|
|
chr1_-_120612301
|
0.149
|
NM_001200001
NM_024408
|
NOTCH2
|
notch 2
|
|
chr2_-_136743037
|
0.149
|
|
DARS
|
aspartyl-tRNA synthetase
|
|
chr22_+_32340592
|
0.148
|
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide
|
|
chr1_-_33336289
|
0.148
|
NM_001171940
NM_153756
|
FNDC5
|
fibronectin type III domain containing 5
|
|
chr2_+_150187028
|
0.146
|
NM_194317
|
LYPD6
|
LY6/PLAUR domain containing 6
|
|
chr1_+_206643786
|
0.146
|
|
IKBKE
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon
|
|
chr18_+_21719447
|
0.146
|
NM_138644
|
CABYR
|
calcium binding tyrosine-(Y)-phosphorylation regulated
|
|
chr22_+_31518923
|
0.145
|
NM_001002837
|
INPP5J
|
inositol polyphosphate-5-phosphatase J
|
|
chr1_-_38273823
|
0.144
|
|
YRDC
|
yrdC domain containing (E. coli)
|
|
chr8_+_31497267
|
0.143
|
NM_013962
|
NRG1
|
neuregulin 1
|
|
chr3_-_187454247
|
0.142
|
NM_001130845
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr21_+_27011766
|
0.142
|
|
JAM2
|
junctional adhesion molecule 2
|
|
chr17_+_7255207
|
0.142
|
NM_001002914
|
KCTD11
|
potassium channel tetramerisation domain containing 11
|
|
chr11_+_7273141
|
0.142
|
NM_175733
|
SYT9
|
synaptotagmin IX
|
|
chr3_-_158450230
|
0.141
|
|
RARRES1
|
retinoic acid receptor responder (tazarotene induced) 1
|
|
chr11_-_65667858
|
0.140
|
|
FOSL1
|
FOS-like antigen 1
|
|
chr2_-_131850988
|
0.140
|
NM_001009993
|
FAM168B
|
family with sequence similarity 168, member B
|
|
chr1_-_12677347
|
0.139
|
NM_004753
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3
|
|
chr1_-_894651
|
0.139
|
NM_015658
|
NOC2L
|
nucleolar complex associated 2 homolog (S. cerevisiae)
|
|
chr7_-_142583260
|
0.139
|
NM_018646
|
TRPV6
|
transient receptor potential cation channel, subfamily V, member 6
|
|
chr10_+_99400469
|
0.139
|
|
PI4K2A
|
phosphatidylinositol 4-kinase type 2 alpha
|
|
chr14_+_24630421
|
0.138
|
NM_006084
|
IRF9
|
interferon regulatory factor 9
|
|
chr16_-_69373406
|
0.138
|
|
COG8
|
component of oligomeric golgi complex 8
|
|
chr7_-_2354010
|
0.138
|
|
SNX8
|
sorting nexin 8
|
|
chr7_+_108210188
|
0.137
|
NM_012328
|
DNAJB9
|
DnaJ (Hsp40) homolog, subfamily B, member 9
|
|
chr18_+_21718930
|
0.137
|
NM_012189
NM_138643
NM_153768
NM_153769
NM_153770
|
CABYR
|
calcium binding tyrosine-(Y)-phosphorylation regulated
|
|
chr2_-_239197091
|
0.137
|
NM_022817
|
PER2
|
period homolog 2 (Drosophila)
|
|
chr6_+_44095357
|
0.137
|
NM_018426
|
TMEM63B
|
transmembrane protein 63B
|
|
chr3_+_5021096
|
0.137
|
NM_003670
|
BHLHE40
|
basic helix-loop-helix family, member e40
|
|
chr19_+_38794800
|
0.137
|
NM_033520
|
C19orf33
|
chromosome 19 open reading frame 33
|
|
chr1_+_154975041
|
0.137
|
NM_001252405
NM_015872
|
ZBTB7B
|
zinc finger and BTB domain containing 7B
|
|
chr1_+_170501240
|
0.136
|
NM_001146039
NM_152281
|
GORAB
|
golgin, RAB6-interacting
|
|
chr6_+_29974352
|
0.136
|
|
HLA-J
|
major histocompatibility complex, class I, J (pseudogene)
|
|
chr22_+_32750871
|
0.136
|
NM_006604
|
RFPL3
|
ret finger protein-like 3
|
|
chr11_+_35160666
|
0.136
|
|
CD44
|
CD44 molecule (Indian blood group)
|
|
chr1_+_156084579
|
0.136
|
|
LMNA
|
lamin A/C
|
|
chr16_-_86542463
|
0.134
|
|
LOC400550
|
uncharacterized LOC400550
|
|
chr15_+_74287132
|
0.134
|
|
PML
|
promyelocytic leukemia
|
|
chr19_-_1174257
|
0.134
|
|
SBNO2
|
strawberry notch homolog 2 (Drosophila)
|
|
chr21_-_35899046
|
0.133
|
NM_203418
|
RCAN1
|
regulator of calcineurin 1
|
|
chr2_+_54683276
|
0.133
|
NM_003128
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1
|
|
chrX_+_70315637
|
0.132
|
NM_001170931
NM_005938
|
FOXO4
|
forkhead box O4
|
|
chr17_+_41561232
|
0.132
|
NM_004941
|
DHX8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8
|
|
chr10_+_99400441
|
0.132
|
NM_018425
|
PI4K2A
|
phosphatidylinositol 4-kinase type 2 alpha
|