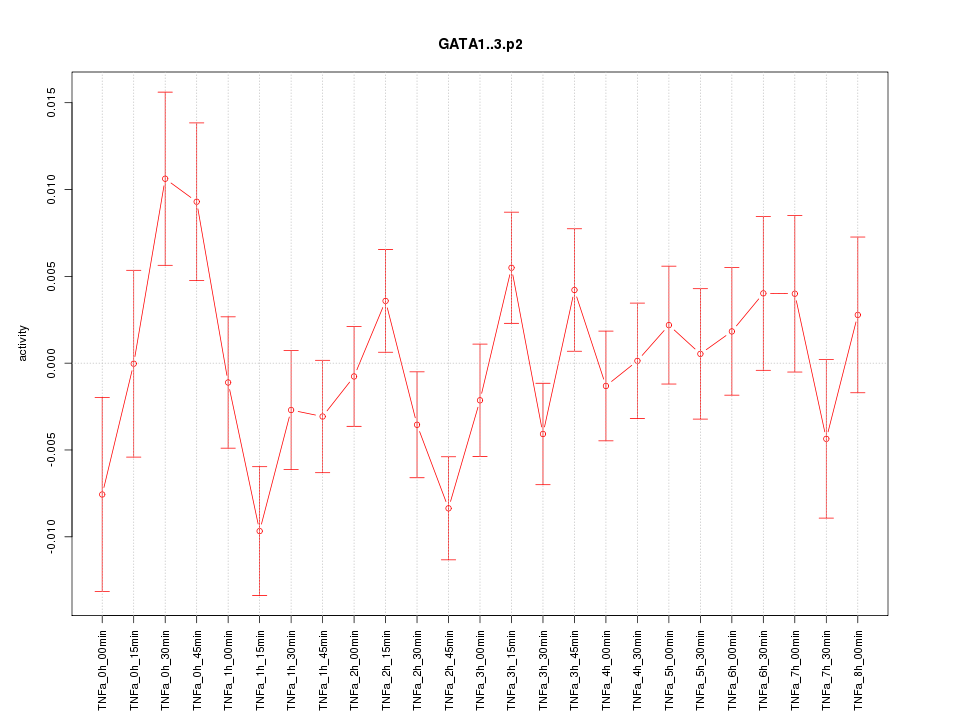
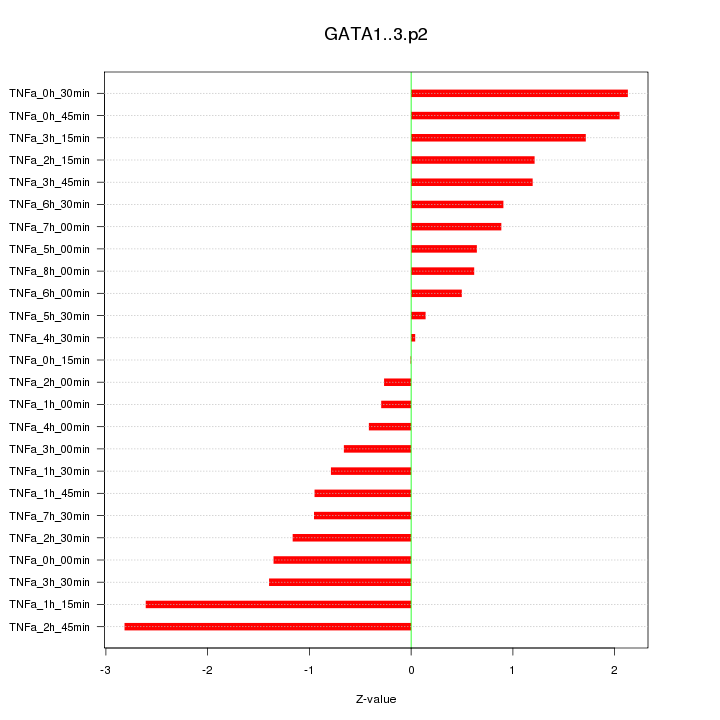
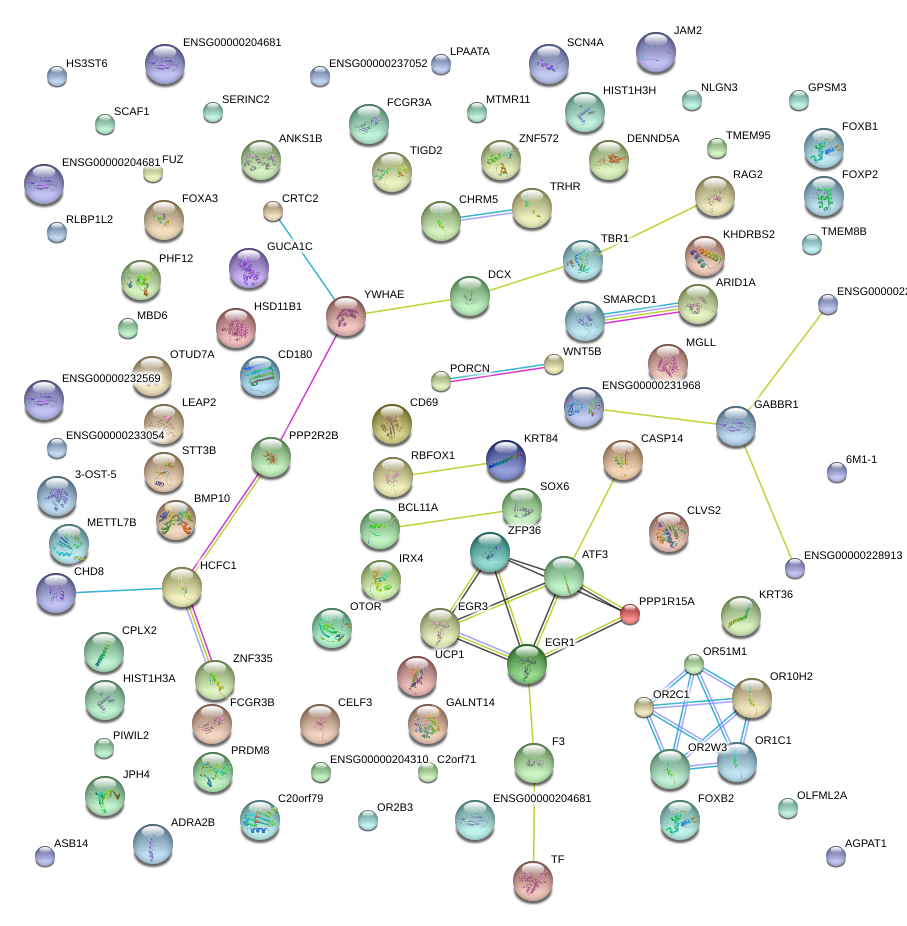
|
chr8_-_22550057
|
1.692
|
NM_001199881
|
EGR3
|
early growth response 3
|
|
chr11_+_5410606
|
1.394
|
NM_001004756
|
OR51M1
|
olfactory receptor, family 51, subfamily M, member 1
|
|
chr12_-_52779362
|
1.268
|
NM_033045
|
KRT84
|
keratin 84
|
|
chr6_-_114384008
|
1.168
|
NM_153612
|
HS3ST5
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 5
|
|
chr16_+_3405888
|
1.143
|
NM_012368
|
OR2C1
|
olfactory receptor, family 2, subfamily C, member 1
|
|
chr19_+_15160194
|
1.051
|
NM_012114
|
CASP14
|
caspase 14, apoptosis-related cysteine peptidase
|
|
chr9_+_35829375
|
1.050
|
NM_001042590
|
TMEM8B
|
transmembrane protein 8B
|
|
chrX_-_153236621
|
0.984
|
NM_005334
|
HCFC1
|
host cell factor C1 (VP16-accessory protein)
|
|
chr2_-_60780767
|
0.896
|
|
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein)
|
|
chr19_+_50145369
|
0.894
|
NM_021228
|
SCAF1
|
SR-related CTD-associated factor 1
|
|
chr2_+_162272892
|
0.884
|
|
TBR1
|
T-box, brain, 1
|
|
chr8_-_22550708
|
0.836
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr3_+_133494275
|
0.830
|
|
TF
|
transferrin
|
|
chr22_+_22385340
|
0.817
|
|
|
|
|
chr19_+_49376488
|
0.802
|
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr17_-_62050200
|
0.795
|
NM_000334
|
SCN4A
|
sodium channel, voltage-gated, type IV, alpha subunit
|
|
chr8_+_110099652
|
0.787
|
NM_003301
|
TRHR
|
thyrotropin-releasing hormone receptor
|
|
chr8_-_22549901
|
0.784
|
NM_001199880
|
EGR3
|
early growth response 3
|
|
chr2_-_31352489
|
0.771
|
NM_001253827
|
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14)
|
|
chr16_+_89778242
|
0.751
|
|
LOC100128881
|
uncharacterized LOC100128881
|
|
chr19_+_39897457
|
0.737
|
NM_003407
|
ZFP36
|
zinc finger protein 36, C3H type, homolog (mouse)
|
|
chr8_+_22210311
|
0.711
|
|
PIWIL2
|
piwi-like 2 (Drosophila)
|
|
chr11_-_36619776
|
0.707
|
NM_000536
NM_001243785
NM_001243786
|
RAG2
|
recombination activating gene 2
|
|
chr1_-_153931077
|
0.703
|
|
CRTC2
|
CREB regulated transcription coactivator 2
|
|
chr11_-_9225780
|
0.688
|
|
DENND5A
|
DENN/MADD domain containing 5A
|
|
chr3_-_127541160
|
0.683
|
NM_007283
|
MGLL
|
monoglyceride lipase
|
|
chr6_-_29595746
|
0.678
|
|
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1
|
|
chr5_+_132209354
|
0.668
|
NM_052971
|
LEAP2
|
liver expressed antimicrobial peptide 2
|
|
chr2_-_69098502
|
0.663
|
NM_014482
|
BMP10
|
bone morphogenetic protein 10
|
|
chr9_+_35829208
|
0.648
|
NM_016446
NM_001042589
|
TMEM8B
|
transmembrane protein 8B
|
|
chr11_-_64377332
|
0.646
|
|
|
|
|
chr1_+_27101217
|
0.629
|
|
ARID1A
|
AT rich interactive domain 1A (SWI-like)
|
|
chr11_-_16424391
|
0.628
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr1_+_212738675
|
0.624
|
NM_001030287
|
ATF3
|
activating transcription factor 3
|
|
chr1_-_153930911
|
0.618
|
|
CRTC2
|
CREB regulated transcription coactivator 2
|
|
chr5_+_137801166
|
0.607
|
|
EGR1
|
early growth response 1
|
|
chr17_+_7258454
|
0.601
|
NM_198154
|
TMEM95
|
transmembrane protein 95
|
|
chr20_+_16728997
|
0.594
|
NM_020157
|
OTOR
|
otoraplin
|
|
chr5_-_66492574
|
0.584
|
NM_005582
|
CD180
|
CD180 molecule
|
|
chr1_+_248058857
|
0.582
|
NM_001001957
|
OR2W3
|
olfactory receptor, family 2, subfamily W, member 3
|
|
chr17_-_1303402
|
0.579
|
|
YWHAE
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide
|
|
chr20_-_44600939
|
0.578
|
|
ZNF335
|
zinc finger protein 335
|
|
chr1_-_151688791
|
0.568
|
NM_001172648
NM_001172649
|
CELF3
|
CUGBP, Elav-like family member 3
|
|
chr3_-_57312979
|
0.554
|
NM_130387
|
ASB14
|
ankyrin repeat and SOCS box containing 14
|
|
chr6_+_123317377
|
0.541
|
|
CLVS2
|
clavesin 2
|
|
chr17_-_39646115
|
0.537
|
NM_003771
|
KRT36
|
keratin 36
|
|
chr6_+_123317579
|
0.534
|
NM_001010852
|
CLVS2
|
clavesin 2
|
|
chr3_+_31574431
|
0.533
|
|
STT3B
|
STT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae)
|
|
chr12_+_57920425
|
0.531
|
|
MBD6
|
methyl-CpG binding domain protein 6
|
|
chr4_-_141489882
|
0.528
|
NM_021833
|
UCP1
|
uncoupling protein 1 (mitochondrial, proton carrier)
|
|
chr16_+_6533758
|
0.528
|
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr15_+_60296420
|
0.527
|
NM_012182
|
FOXB1
|
forkhead box B1
|
|
chr1_-_161601251
|
0.527
|
NM_001244753
|
FCGR3B
|
Fc fragment of IgG, low affinity IIIb, receptor (CD16b)
|
|
chr1_+_31882411
|
0.526
|
NM_001199038
|
SERINC2
|
serine incorporator 2
|
|
chr8_+_125985538
|
0.525
|
NM_152412
|
ZNF572
|
zinc finger protein 572
|
|
chr12_-_9913415
|
0.523
|
NM_001781
|
CD69
|
CD69 molecule
|
|
chr12_+_50479033
|
0.522
|
|
SMARCD1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1
|
|
chr2_-_29297126
|
0.516
|
NM_001029883
|
C2orf71
|
chromosome 2 open reading frame 71
|
|
chr1_-_161519817
|
0.515
|
NM_000569
NM_001127592
|
FCGR3A
|
Fc fragment of IgG, low affinity IIIa, receptor (CD16a)
|
|
chr14_-_21899866
|
0.514
|
NM_001170629
|
CHD8
|
chromodomain helicase DNA binding protein 8
|
|
chr7_+_114055171
|
0.512
|
|
FOXP2
|
forkhead box P2
|
|
chr19_-_50316360
|
0.511
|
NM_001171937
NM_025129
|
FUZ
|
fuzzy homolog (Drosophila)
|
|
chr15_-_31947541
|
0.509
|
NM_130901
|
OTUD7A
|
OTU domain containing 7A
|
|
chr15_+_34261088
|
0.509
|
NM_012125
|
CHRM5
|
cholinergic receptor, muscarinic 5
|
|
chr5_+_137801178
|
0.507
|
NM_001964
|
EGR1
|
early growth response 1
|
|
chr14_-_24047428
|
0.507
|
|
JPH4
|
junctophilin 4
|
|
chr2_-_96781887
|
0.507
|
NM_000682
|
ADRA2B
|
adrenergic, alpha-2B-, receptor
|
|
chr9_+_127420690
|
0.505
|
|
MIR181A2HG
|
MIR181A2 host gene (non-protein coding)
|
|
chr6_-_32143795
|
0.504
|
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha)
|
|
chr4_+_90033967
|
0.499
|
NM_145715
|
TIGD2
|
tigger transposable element derived 2
|
|
chr3_+_31574119
|
0.497
|
|
STT3B
|
STT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae)
|
|
chr12_+_56075418
|
0.496
|
|
METTL7B
|
methyltransferase like 7B
|
|
chr17_-_48271365
|
0.496
|
|
|
|
|
chr3_-_108672547
|
0.495
|
NM_005459
|
GUCA1C
|
guanylate cyclase activator 1C
|
|
chr12_-_122240827
|
0.492
|
|
|
|
|
chr5_-_146435589
|
0.489
|
NM_181674
NM_181676
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta
|
|
chr6_-_29055048
|
0.488
|
NM_001005226
|
OR2B3
|
olfactory receptor, family 2, subfamily B, member 3
|
|
chr19_+_15838833
|
0.483
|
NM_013939
|
OR10H2
|
olfactory receptor, family 10, subfamily H, member 2
|
|
chr6_+_27777713
|
0.482
|
NM_003536
|
HIST1H3H
|
histone cluster 1, H3h
|
|
chr21_+_27012067
|
0.480
|
|
JAM2
|
junctional adhesion molecule 2
|
|
chr20_+_18794369
|
0.479
|
NM_178483
|
C20orf79
|
chromosome 20 open reading frame 79
|
|
chr1_+_212788359
|
0.475
|
NM_001206486
|
ATF3
|
activating transcription factor 3
|
|
chrX_+_70364680
|
0.473
|
NM_001166660
NM_018977
NM_181303
|
NLGN3
|
neuroligin 3
|
|
chr17_-_27240982
|
0.472
|
|
PHF12
|
PHD finger protein 12
|
|
chrX_+_48368063
|
0.472
|
NM_203474
NM_203475
|
PORCN
|
porcupine homolog (Drosophila)
|
|
chr12_-_99548471
|
0.471
|
NM_001204068
NM_001204069
NM_001204070
NM_020140
NM_181670
NM_001204066
|
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr18_+_77659446
|
0.469
|
|
|
|
|
chr1_-_151689246
|
0.466
|
NM_007185
|
CELF3
|
CUGBP, Elav-like family member 3
|
|
chr12_+_1738411
|
0.465
|
NM_032642
|
WNT5B
|
wingless-type MMTV integration site family, member 5B
|
|
chr6_-_32160289
|
0.464
|
|
GPSM3
|
G-protein signaling modulator 3
|
|
chr5_-_1882764
|
0.463
|
NM_016358
|
IRX4
|
iroquois homeobox 4
|
|
chr1_-_149908759
|
0.461
|
NM_001145862
|
MTMR11
|
myotubularin related protein 11
|
|
chr1_-_153931096
|
0.460
|
NM_181715
|
CRTC2
|
CREB regulated transcription coactivator 2
|
|
chr1_+_209878135
|
0.460
|
NM_005525
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1
|
|
chr19_+_46367517
|
0.459
|
NM_004497
|
FOXA3
|
forkhead box A3
|
|
chr5_+_175308843
|
0.459
|
|
CPLX2
|
complexin 2
|
|
chrX_-_110654373
|
0.459
|
NM_000555
|
DCX
|
doublecortin
|
|
chr1_-_247921707
|
0.458
|
NM_012353
|
OR1C1
|
olfactory receptor, family 1, subfamily C, member 1
|
|
chr6_-_32143886
|
0.458
|
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha)
|
|
chr4_+_81118639
|
0.457
|
NM_001099403
|
PRDM8
|
PR domain containing 8
|
|
chr6_-_62995852
|
0.457
|
NM_152688
|
KHDRBS2
|
KH domain containing, RNA binding, signal transduction associated 2
|
|
chr13_-_33780142
|
0.455
|
NM_001243474
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr7_+_103969103
|
0.453
|
NM_199000
|
LHFPL3
|
lipoma HMGIC fusion partner-like 3
|
|
chr4_-_21305528
|
0.453
|
NM_147183
|
KCNIP4
|
Kv channel interacting protein 4
|
|
chr11_-_72385411
|
0.451
|
NM_002599
|
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated
|
|
chr7_+_39017608
|
0.449
|
NM_001166018
NM_007252
|
POU6F2
|
POU class 6 homeobox 2
|
|
chr1_-_26644755
|
0.447
|
NM_145345
|
UBXN11
|
UBX domain protein 11
|
|
chr17_-_1248346
|
0.444
|
|
PAFAH1B1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa)
|
|
chr17_-_39150384
|
0.444
|
NM_033185
|
KRTAP3-3
|
keratin associated protein 3-3
|
|
chr1_+_47137496
|
0.442
|
NM_001145474
|
ATPAF1-AS1
|
ATPAF1 antisense RNA 1 (non-protein coding)
|
|
chr16_+_30709913
|
0.440
|
|
|
|
|
chrX_+_70364707
|
0.437
|
|
NLGN3
|
neuroligin 3
|
|
chr16_+_24802421
|
0.435
|
|
TNRC6A
|
trinucleotide repeat containing 6A
|
|
chr11_-_70858241
|
0.435
|
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr10_-_100995558
|
0.434
|
NM_001166244
NM_001166245
NM_001166246
NM_021828
|
HPSE2
|
heparanase 2
|
|
chr17_-_39280377
|
0.434
|
NM_031854
|
KRTAP4-12
|
keratin associated protein 4-12
|
|
chr6_-_32143899
|
0.431
|
NM_006411
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha)
|
|
chr8_+_102504836
|
0.431
|
|
GRHL2
|
grainyhead-like 2 (Drosophila)
|
|
chr1_+_153232178
|
0.427
|
NM_000427
|
LOR
|
loricrin
|
|
chr15_-_72523542
|
0.425
|
|
PKM2
|
pyruvate kinase, muscle
|
|
chr4_+_69681709
|
0.425
|
NM_001075
NM_001144767
|
UGT2B10
|
UDP glucuronosyltransferase 2 family, polypeptide B10
|
|
chr12_+_94542239
|
0.425
|
NM_005761
|
PLXNC1
|
plexin C1
|
|
chr5_-_172755418
|
0.423
|
|
STC2
|
stanniocalcin 2
|
|
chrX_+_70364728
|
0.421
|
|
NLGN3
|
neuroligin 3
|
|
chrX_+_82763268
|
0.420
|
NM_000307
|
POU3F4
|
POU class 3 homeobox 4
|
|
chrX_+_49126480
|
0.419
|
|
PPP1R3F
|
protein phosphatase 1, regulatory subunit 3F
|
|
chr19_-_51141301
|
0.414
|
NM_032298
|
SYT3
|
synaptotagmin III
|
|
chr16_+_67694795
|
0.413
|
NM_001037281
NM_016948
|
PARD6A
|
par-6 partitioning defective 6 homolog alpha (C. elegans)
|
|
chr20_-_1309809
|
0.411
|
NM_080489
|
SDCBP2
|
syndecan binding protein (syntenin) 2
|
|
chr6_-_119470336
|
0.410
|
NM_001100411
|
FAM184A
|
family with sequence similarity 184, member A
|
|
chr2_+_169757749
|
0.410
|
NM_001081686
NM_021176
|
G6PC2
|
glucose-6-phosphatase, catalytic, 2
|
|
chr15_-_72523483
|
0.409
|
|
PKM2
|
pyruvate kinase, muscle
|
|
chr9_+_131191206
|
0.408
|
|
CERCAM
|
cerebral endothelial cell adhesion molecule
|
|
chr9_-_34458477
|
0.408
|
NM_001184940
NM_001184941
NM_001184942
NM_001184943
NM_001184945
NM_147202
|
C9orf25
|
chromosome 9 open reading frame 25
|
|
chr15_-_72523557
|
0.407
|
|
PKM2
|
pyruvate kinase, muscle
|
|
chr12_+_7033625
|
0.405
|
NM_001007026
|
ATN1
|
atrophin 1
|
|
chr17_+_42925278
|
0.404
|
NM_016438
|
HIGD1B
|
HIG1 hypoxia inducible domain family, member 1B
|
|
chr5_+_156820161
|
0.402
|
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr5_-_149669187
|
0.400
|
|
CAMK2A
|
calcium/calmodulin-dependent protein kinase II alpha
|
|
chr16_+_29674564
|
0.398
|
|
SPN
|
sialophorin
|
|
chr12_+_81101407
|
0.397
|
NM_002469
|
MYF6
|
myogenic factor 6 (herculin)
|
|
chr18_+_55102777
|
0.397
|
NM_004852
|
ONECUT2
|
one cut homeobox 2
|
|
chr16_+_202853
|
0.397
|
NM_005332
|
HBZ
|
hemoglobin, zeta
|
|
chr16_-_28518131
|
0.396
|
NM_145659
|
IL27
|
interleukin 27
|
|
chr12_+_50478942
|
0.389
|
NM_003076
NM_139071
|
SMARCD1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1
|
|
chr5_-_141993691
|
0.389
|
NM_033137
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr6_-_134639195
|
0.388
|
NM_001143676
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr17_+_36861857
|
0.387
|
NM_005937
|
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6
|
|
chr8_+_50824596
|
0.385
|
NM_018967
|
SNTG1
|
syntrophin, gamma 1
|
|
chr5_+_31193852
|
0.383
|
|
CDH6
|
cadherin 6, type 2, K-cadherin (fetal kidney)
|
|
chr1_-_145715561
|
0.379
|
NM_007053
|
CD160
|
CD160 molecule
|
|
chr2_+_36923832
|
0.379
|
NM_001177969
NM_001177970
NM_001177971
NM_001177972
NM_053276
|
VIT
|
vitrin
|
|
chr5_-_145895675
|
0.378
|
NM_194251
|
GPR151
|
G protein-coupled receptor 151
|
|
chr19_-_46272496
|
0.378
|
NM_175875
|
SIX5
|
SIX homeobox 5
|
|
chr8_+_65492794
|
0.377
|
NM_152414
|
BHLHE22
|
basic helix-loop-helix family, member e22
|
|
chr11_-_5173598
|
0.374
|
NM_012375
|
OR52A1
|
olfactory receptor, family 52, subfamily A, member 1
|
|
chr8_+_145064583
|
0.373
|
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding)
|
|
chr9_+_18474078
|
0.372
|
NM_001040272
NM_052866
|
ADAMTSL1
|
ADAMTS-like 1
|
|
chr11_-_47399903
|
0.371
|
|
|
|
|
chr13_+_112721912
|
0.369
|
NM_005986
|
SOX1
|
SRY (sex determining region Y)-box 1
|
|
chr11_+_69455938
|
0.368
|
|
CCND1
|
cyclin D1
|
|
chr9_-_35115844
|
0.367
|
NM_025182
|
FAM214B
|
family with sequence similarity 214, member B
|
|
chr16_+_23847321
|
0.366
|
|
PRKCB
|
protein kinase C, beta
|
|
chr5_-_172198160
|
0.366
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chr2_-_51259239
|
0.366
|
|
NRXN1
|
neurexin 1
|
|
chr19_+_15197790
|
0.365
|
NM_001004713
|
OR1I1
|
olfactory receptor, family 1, subfamily I, member 1
|
|
chr13_-_33859835
|
0.362
|
NM_178006
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr7_-_31380507
|
0.361
|
NM_022728
|
NEUROD6
|
neurogenic differentiation 6
|
|
chr16_-_67881360
|
0.357
|
NM_025082
|
CENPT
|
centromere protein T
|
|
chr2_-_74734498
|
0.355
|
|
PCGF1
|
polycomb group ring finger 1
|
|
chr15_+_60296506
|
0.354
|
|
FOXB1
|
forkhead box B1
|
|
chrX_+_128913897
|
0.353
|
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chr11_-_120008219
|
0.353
|
|
TRIM29
|
tripartite motif containing 29
|
|
chr19_-_14224979
|
0.352
|
NM_207518
|
PRKACA
|
protein kinase, cAMP-dependent, catalytic, alpha
|
|
chr2_-_89619820
|
0.350
|
|
IGKC
|
immunoglobulin kappa constant
|
|
chr10_+_85933553
|
0.350
|
NM_207373
|
C10orf99
|
chromosome 10 open reading frame 99
|
|
chr1_-_111743261
|
0.349
|
NM_024901
|
DENND2D
|
DENN/MADD domain containing 2D
|
|
chr6_+_43265994
|
0.349
|
NM_006672
NM_153320
|
SLC22A7
|
solute carrier family 22 (organic anion transporter), member 7
|
|
chr17_-_48271968
|
0.348
|
|
|
|
|
chr16_-_27561216
|
0.347
|
|
GTF3C1
|
general transcription factor IIIC, polypeptide 1, alpha 220kDa
|
|
chr19_+_50094911
|
0.346
|
NM_020719
|
PRR12
|
proline rich 12
|
|
chr18_-_28742744
|
0.345
|
NM_004948
NM_024421
|
DSC1
|
desmocollin 1
|
|
chr11_-_5345530
|
0.345
|
NM_033180
|
OR51B2
|
olfactory receptor, family 51, subfamily B, member 2
|
|
chr19_-_46295647
|
0.341
|
|
DMWD
|
dystrophia myotonica, WD repeat containing
|
|
chr6_+_31543340
|
0.341
|
NM_000594
|
TNF
|
tumor necrosis factor
|
|
chr12_-_99548867
|
0.341
|
NM_001204067
|
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr12_-_123380628
|
0.339
|
|
VPS37B
|
vacuolar protein sorting 37 homolog B (S. cerevisiae)
|
|
chr8_-_121824015
|
0.338
|
|
|
|
|
chr4_+_76481257
|
0.338
|
NM_001206981
NM_178497
|
C4orf26
|
chromosome 4 open reading frame 26
|
|
chr17_-_1303548
|
0.337
|
NM_006761
|
YWHAE
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide
|
|
chr22_+_31478845
|
0.333
|
NM_001207017
|
SMTN
|
smoothelin
|
|
chr9_+_35560424
|
0.332
|
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr9_-_35958095
|
0.332
|
NM_019897
|
OR2S2
|
olfactory receptor, family 2, subfamily S, member 2
|
|
chr19_-_54692132
|
0.331
|
|
MBOAT7
|
membrane bound O-acyltransferase domain containing 7
|
|
chr7_+_141463896
|
0.329
|
NM_016943
|
TAS2R3
|
taste receptor, type 2, member 3
|
|
chr20_-_44600787
|
0.328
|
NM_022095
|
ZNF335
|
zinc finger protein 335
|
|
chr20_+_327329
|
0.326
|
NM_024958
|
NRSN2
|
neurensin 2
|
|
chrX_+_41306686
|
0.324
|
NM_022567
|
NYX
|
nyctalopin
|
|
chr2_-_60780404
|
0.324
|
NM_018014
NM_022893
NM_138559
|
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein)
|
|
chr11_-_1017140
|
0.323
|
|
MUC6
|
mucin 6, oligomeric mucus/gel-forming
|