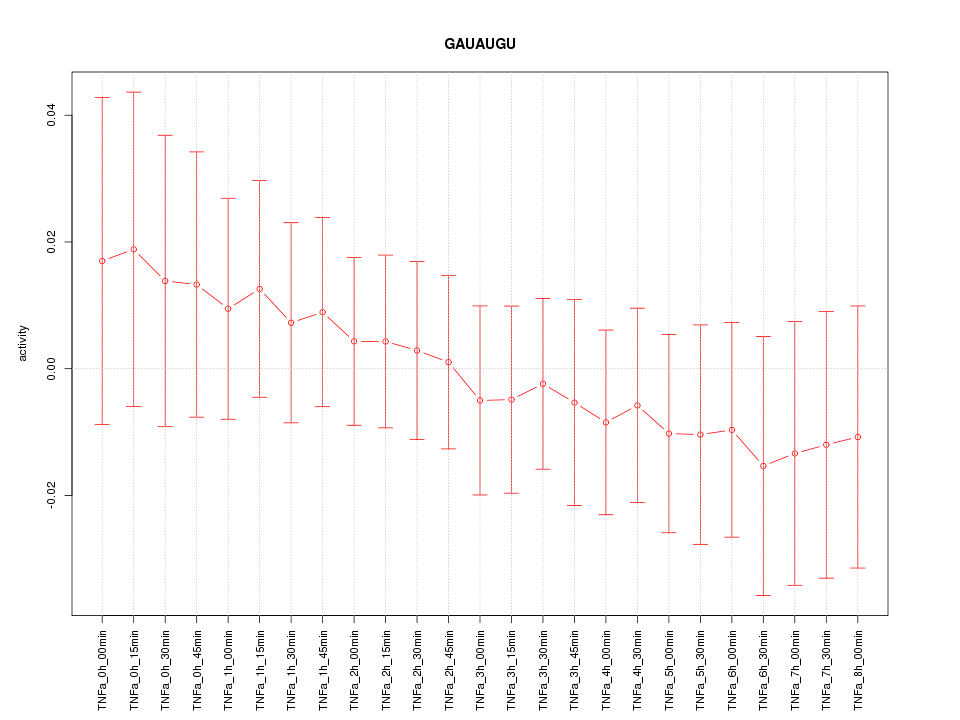
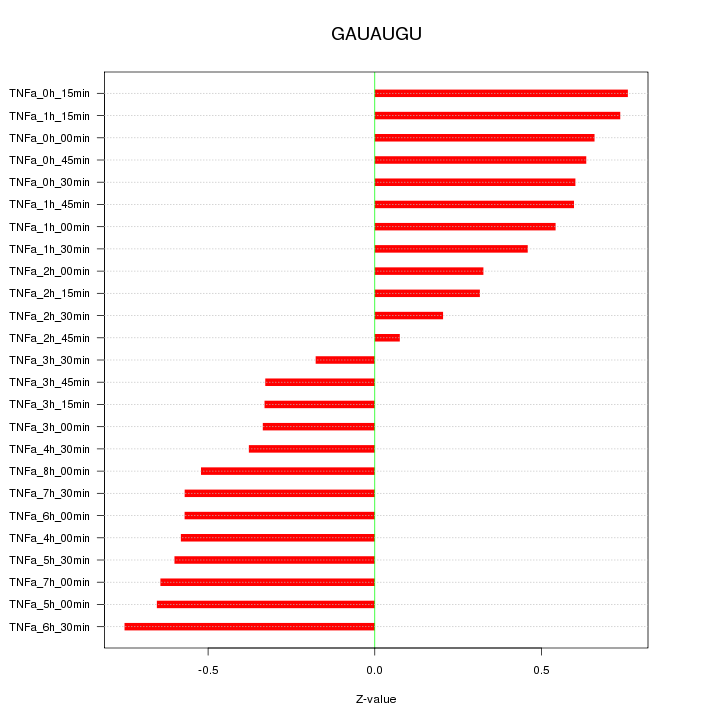
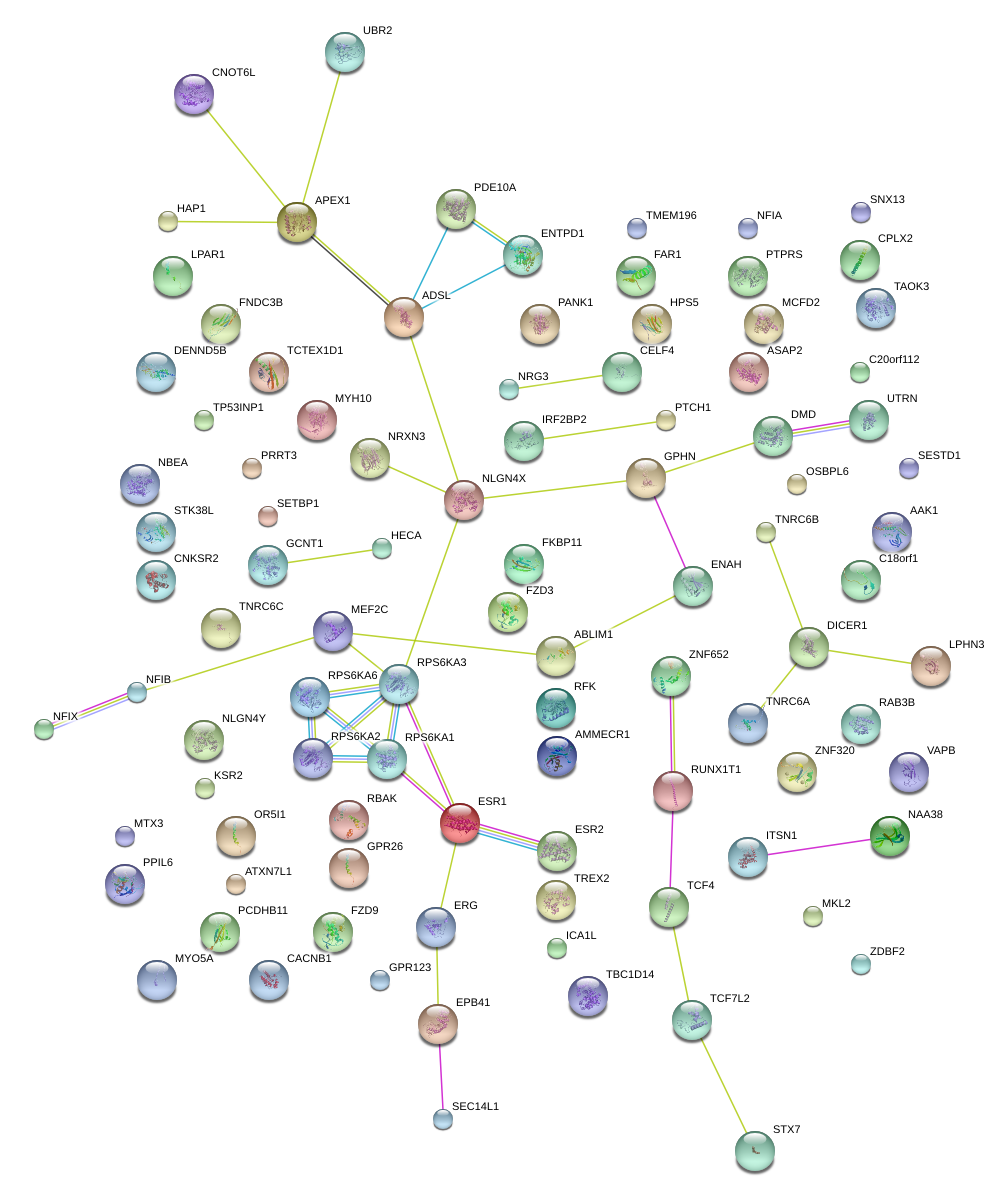
|
chr12_+_27397077
|
1.600
|
NM_015000
|
STK38L
|
serine/threonine kinase 38 like
|
|
chr9_-_14398981
|
0.896
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chr1_+_61542928
|
0.842
|
NM_001145511
|
NFIA
|
nuclear factor I/A
|
|
chr8_-_93029864
|
0.829
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr9_-_14314036
|
0.820
|
NM_001190737
NM_005596
|
NFIB
|
nuclear factor I/B
|
|
chr1_+_61547625
|
0.815
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr12_-_31743822
|
0.796
|
NM_144973
|
DENND5B
|
DENN/MADD domain containing 5B
|
|
chr8_-_93107881
|
0.781
|
NM_001198625
NM_001198626
NM_001198627
NM_001198631
NM_001198634
NM_001198679
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_-_93111915
|
0.769
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr1_+_61547388
|
0.761
|
NM_001145512
|
NFIA
|
nuclear factor I/A
|
|
chr8_-_93107442
|
0.743
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_-_93115453
|
0.685
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_-_93075190
|
0.616
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr9_+_79056581
|
0.611
|
NM_001097634
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chr18_+_42260774
|
0.589
|
NM_015559
|
SETBP1
|
SET binding protein 1
|
|
chr9_+_79093256
|
0.571
|
NM_001097635
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chr9_+_79115548
|
0.568
|
NM_001097636
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chr9_+_79074067
|
0.566
|
NM_001097633
NM_001490
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chr13_+_35516390
|
0.501
|
NM_015678
|
NBEA
|
neurobeachin
|
|
chr14_+_66974124
|
0.466
|
NM_001024218
NM_020806
|
GPHN
|
gephyrin
|
|
chr5_-_88179278
|
0.443
|
NM_001193350
NM_002397
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr13_+_36050885
|
0.435
|
NM_001204197
|
NBEA
|
neurobeachin
|
|
chr2_+_179184970
|
0.435
|
NM_145739
|
OSBPL6
|
oxysterol binding protein-like 6
|
|
chr2_+_179059071
|
0.429
|
NM_001201480
NM_001201481
NM_001201482
NM_032523
|
OSBPL6
|
oxysterol binding protein-like 6
|
|
chr5_-_88199868
|
0.411
|
NM_001131005
NM_001193347
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr17_-_8534008
|
0.408
|
NM_005964
|
MYH10
|
myosin, heavy chain 10, non-muscle
|
|
chr10_-_116418056
|
0.374
|
NM_002313
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr10_-_116286661
|
0.373
|
NM_006720
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr16_+_14164879
|
0.372
|
NM_014048
|
MKL2
|
MKL/myocardin-like 2
|
|
chr10_-_116444374
|
0.360
|
NM_001003407
NM_001003408
|
ABLIM1
|
actin binding LIM protein 1
|
|
chrX_-_33229428
|
0.349
|
NM_004006
|
DMD
|
dystrophin
|
|
chr5_-_88119604
|
0.327
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chrX_-_32173585
|
0.317
|
NM_004013
NM_004020
NM_004021
NM_004022
NM_004023
|
DMD
|
dystrophin
|
|
chr17_+_76000248
|
0.308
|
NM_001142640
NM_018996
|
TNRC6C
|
trinucleotide repeat containing 6C
|
|
chrX_-_33146543
|
0.292
|
NM_004010
NM_004009
|
DMD
|
dystrophin
|
|
chr18_-_53255734
|
0.290
|
NM_001083962
NM_001243230
NM_003199
|
TCF4
|
transcription factor 4
|
|
chrX_-_32430370
|
0.273
|
NM_004011
NM_004012
|
DMD
|
dystrophin
|
|
chr1_-_234745223
|
0.267
|
NM_001077397
NM_182972
|
IRF2BP2
|
interferon regulatory factor 2 binding protein 2
|
|
chrX_-_31526414
|
0.255
|
NM_004014
|
DMD
|
dystrophin
|
|
chr2_+_9346876
|
0.240
|
NM_001135191
NM_003887
|
ASAP2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2
|
|
chrX_-_31284946
|
0.239
|
NM_004015
NM_004016
NM_004017
NM_004018
NM_004019
|
DMD
|
dystrophin
|
|
chr9_-_98269480
|
0.235
|
NM_001083607
|
PTCH1
|
patched 1
|
|
chr9_-_98270321
|
0.228
|
NM_001083604
NM_001083605
|
PTCH1
|
patched 1
|
|
chr15_-_52821004
|
0.224
|
NM_000259
NM_001142495
|
MYO5A
|
myosin VA (heavy chain 12, myoxin)
|
|
chr9_-_98270830
|
0.211
|
NM_000264
|
PTCH1
|
patched 1
|
|
chrX_-_109561314
|
0.210
|
NM_001025580
NM_015365
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr21_-_40033617
|
0.193
|
NM_001243432
NM_001136154
NM_004449
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chrX_-_33357725
|
0.192
|
NM_000109
|
DMD
|
dystrophin
|
|
chr21_-_39870303
|
0.186
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr9_-_79009265
|
0.170
|
NM_018339
|
RFK
|
riboflavin kinase
|
|
chr5_+_140579182
|
0.170
|
NM_018931
|
PCDHB11
|
protocadherin beta 11
|
|
chr10_+_83637442
|
0.162
|
NM_001165973
|
NRG3
|
neuregulin 3
|
|
chrX_-_20284708
|
0.157
|
NM_004586
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3
|
|
chrX_-_109683457
|
0.155
|
NM_001171689
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr10_+_83635069
|
0.149
|
NM_001010848
NM_001165972
|
NRG3
|
neuregulin 3
|
|
chr7_-_17980102
|
0.143
|
NM_015132
|
SNX13
|
sorting nexin 13
|
|
chr10_+_97471535
|
0.135
|
NM_001098175
|
ENTPD1
|
ectonucleoside triphosphate diphosphohydrolase 1
|
|
chr17_-_47439326
|
0.135
|
NM_001145365
NM_014897
|
ZNF652
|
zinc finger protein 652
|
|
chr9_-_98269686
|
0.135
|
NM_001083606
|
PTCH1
|
patched 1
|
|
chr2_-_69870976
|
0.129
|
NM_014911
|
AAK1
|
AP2 associated kinase 1
|
|
chr17_+_75085234
|
0.126
|
NM_001204410
|
SEC14L1
LINC00338
|
SEC14-like 1 (S. cerevisiae)
long intergenic non-protein coding RNA 338
|
|
chr2_+_207139413
|
0.126
|
NM_020923
|
ZDBF2
|
zinc finger, DBF-type containing 2
|
|
chr19_-_5340700
|
0.124
|
NM_002850
NM_130853
NM_130854
NM_130855
|
PTPRS
|
protein tyrosine phosphatase, receptor type, S
|
|
chr7_-_105319608
|
0.124
|
NM_138495
|
ATXN7L1
|
ataxin 7-like 1
|
|
chr6_-_132834165
|
0.117
|
NM_003569
|
STX7
|
syntaxin 7
|
|
chr17_+_75084724
|
0.115
|
NM_001204408
|
SEC14L1
LINC00338
|
SEC14-like 1 (S. cerevisiae)
long intergenic non-protein coding RNA 338
|
|
chr12_-_118810712
|
0.112
|
NM_016281
|
TAOK3
|
TAO kinase 3
|
|
chr16_+_24740945
|
0.110
|
NM_014494
|
TNRC6A
|
trinucleotide repeat containing 6A
|
|
chr17_+_75136985
|
0.106
|
NM_001039573
NM_001143998
NM_001143999
NM_003003
|
SEC14L1
|
SEC14-like 1 (S. cerevisiae)
|
|
chr18_+_13611664
|
0.104
|
NM_001003674
NM_001003675
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr10_+_134901408
|
0.104
|
NM_001083909
|
GPR123
|
G protein-coupled receptor 123
|
|
chr18_+_13620866
|
0.099
|
NM_004338
NM_181483
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr12_-_118406027
|
0.099
|
NM_173598
|
KSR2
|
kinase suppressor of ras 2
|
|
chr1_-_225840660
|
0.099
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr2_-_180129245
|
0.091
|
NM_178123
|
SESTD1
|
SEC14 and spectrin domains 1
|
|
chr8_-_95961543
|
0.089
|
NM_001135733
NM_033285
|
TP53INP1
|
tumor protein p53 inducible nuclear protein 1
|
|
chr7_+_117824085
|
0.089
|
NM_016200
|
NAA38
|
N(alpha)-acetyltransferase 38, NatC auxiliary subunit
|
|
chr10_+_97515408
|
0.087
|
NM_001164178
NM_001164181
NM_001164179
NM_001164182
NM_001164183
NM_001776
|
ENTPD1
|
ectonucleoside triphosphate diphosphohydrolase 1
|
|
chr17_+_75181294
|
0.085
|
NM_001144001
|
SEC14L1
|
SEC14-like 1 (S. cerevisiae)
|
|
chr10_-_91403630
|
0.082
|
NM_138316
NM_148978
|
PANK1
|
pantothenate kinase 1
|
|
chr18_+_13218777
|
0.082
|
NM_181481
NM_181482
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr22_+_40573879
|
0.072
|
NM_001162501
NM_015088
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr12_+_74931550
|
0.071
|
NM_001136262
|
ATXN7L3B
|
ataxin 7-like 3B
|
|
chr6_+_152128453
|
0.068
|
NM_001122740
|
ESR1
|
estrogen receptor 1
|
|
chr4_-_78740508
|
0.064
|
NM_144571
|
CNOT6L
|
CCR4-NOT transcription complex, subunit 6-like
|
|
chr1_+_29213569
|
0.062
|
NM_001166005
NM_001166007
NM_004437
NM_203342
NM_203343
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr7_+_5085451
|
0.060
|
NM_001204513
NM_001204456
NM_021163
|
RBAK-LOC389458
RBAK
|
RBAK-LOC389458 readthrough
RB-associated KRAB zinc finger
|
|
chr9_-_98279246
|
0.057
|
NM_001083602
NM_001083603
|
PTCH1
|
patched 1
|
|
chr22_+_40440664
|
0.053
|
NM_001024843
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr14_+_78870092
|
0.052
|
NM_004796
|
NRXN3
|
neurexin 3
|
|
chr6_+_139456240
|
0.051
|
NM_016217
|
HECA
|
headcase homolog (Drosophila)
|
|
chr10_-_91405320
|
0.051
|
NM_148977
|
PANK1
|
pantothenate kinase 1
|
|
chr11_+_13690151
|
0.045
|
NM_032228
|
FAR1
|
fatty acyl CoA reductase 1
|
|
chr6_+_152128685
|
0.044
|
NM_000125
|
ESR1
|
estrogen receptor 1
|
|
chr5_+_175298492
|
0.040
|
NM_001008220
|
CPLX2
|
complexin 2
|
|
chrX_-_6146655
|
0.039
|
NM_181332
|
NLGN4X
|
neuroligin 4, X-linked
|
|
chr8_+_28351694
|
0.038
|
NM_017412
NM_145866
|
FZD3
|
frizzled family receptor 3
|
|
chr5_-_79286793
|
0.032
|
NM_001010891
NM_001167741
|
MTX3
|
metaxin 3
|
|
chr6_-_109762373
|
0.031
|
NM_001111298
NM_173672
|
PPIL6
|
peptidylprolyl isomerase (cyclophilin)-like 6
|
|
chr7_-_19812403
|
0.029
|
NM_152774
|
TMEM196
|
transmembrane protein 196
|
|
chr21_+_35014624
|
0.029
|
NM_001001132
NM_003024
|
ITSN1
|
intersectin 1 (SH3 domain protein)
|
|
chr14_-_95608054
|
0.027
|
NM_030621
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr3_+_171758289
|
0.025
|
NM_001135095
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr5_+_175223519
|
0.024
|
NM_006650
|
CPLX2
|
complexin 2
|
|
chr6_-_166075520
|
0.023
|
NM_006661
NM_001130690
|
PDE10A
|
phosphodiesterase 10A
|
|
chr10_+_125425870
|
0.023
|
NM_153442
|
GPR26
|
G protein-coupled receptor 26
|
|
chr1_+_67218104
|
0.015
|
NM_152665
|
TCTEX1D1
|
Tctex1 domain containing 1
|
|
chr4_+_6911094
|
0.013
|
NM_001113361
|
TBC1D14
|
TBC1 domain family, member 14
|
|
chr20_-_31071187
|
0.011
|
NM_080616
|
C20orf112
|
chromosome 20 open reading frame 112
|
|
chr17_-_39890882
|
0.011
|
NM_001079870
NM_001079871
NM_177977
|
HAP1
|
huntingtin-associated protein 1
|
|
chr2_-_47143250
|
0.009
|
NM_001171507
NM_001171510
|
MCFD2
|
multiple coagulation factor deficiency 2
|
|
chr20_+_56964162
|
0.008
|
NM_001195677
NM_004738
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C
|
|
chr18_-_35145999
|
0.008
|
NM_001025087
NM_001025088
NM_001025089
NM_020180
|
CELF4
|
CUGBP, Elav-like family member 4
|
|
chr6_+_42531736
|
0.007
|
NM_001184801
NM_015255
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2
|
|
chr1_-_52456292
|
0.007
|
NM_002867
|
RAB3B
|
RAB3B, member RAS oncogene family
|
|
chr19_-_53394598
|
0.006
|
NM_207333
|
ZNF320
|
zinc finger protein 320
|
|
chr2_-_47142950
|
0.004
|
NM_001171506
NM_001171509
NM_139279
|
MCFD2
|
multiple coagulation factor deficiency 2
|
|
chr2_-_203736223
|
0.004
|
NM_138468
|
ICA1L
|
islet cell autoantigen 1,69kDa-like
|
|
chr6_+_152126807
|
0.002
|
NM_001122741
|
ESR1
|
estrogen receptor 1
|