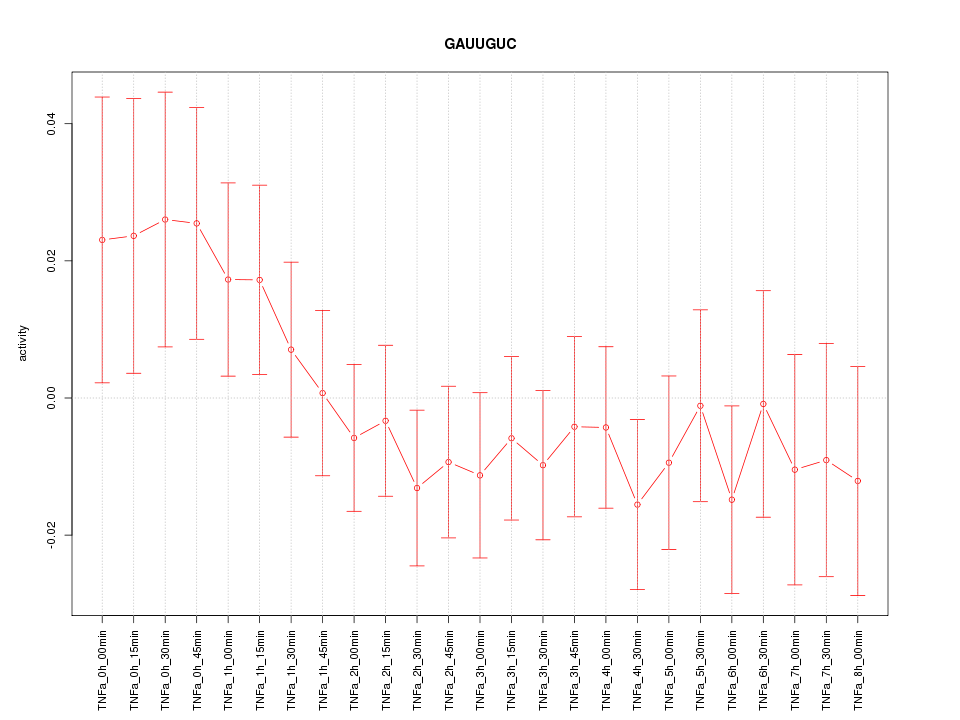
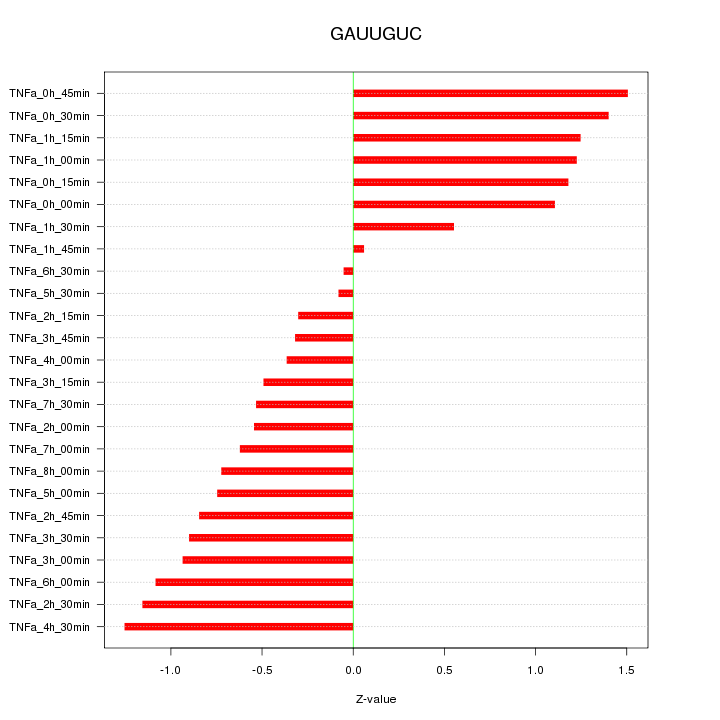
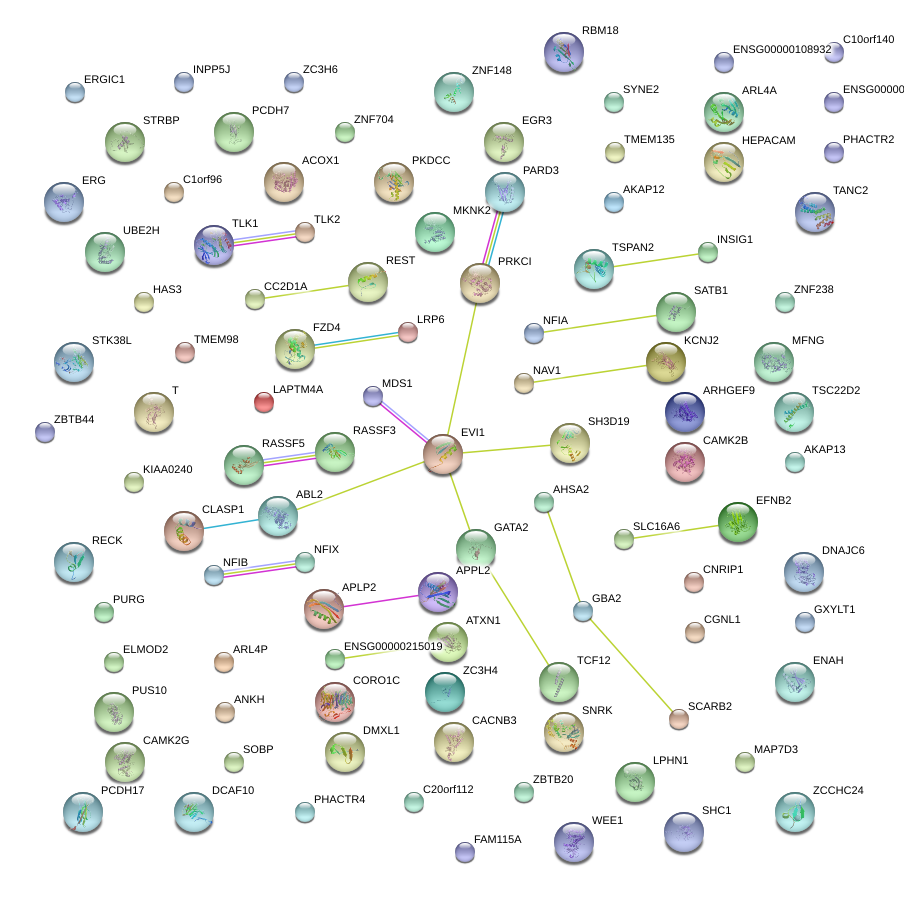
|
chr8_-_22549901
|
4.860
|
NM_001199880
|
EGR3
|
early growth response 3
|
|
chr8_-_22550708
|
4.817
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr8_-_22550057
|
4.733
|
NM_001199881
|
EGR3
|
early growth response 3
|
|
chr13_+_58205788
|
4.304
|
NM_001040429
|
PCDH17
|
protocadherin 17
|
|
chr9_-_35749209
|
3.832
|
NM_020944
|
GBA2
|
glucosidase, beta (bile acid) 2
|
|
chr3_+_43328001
|
3.691
|
NM_001100594
NM_017719
|
SNRK
|
SNF related kinase
|
|
chr10_-_81205091
|
3.326
|
NM_153367
|
ZCCHC24
|
zinc finger, CCHC domain containing 24
|
|
chr15_+_57668702
|
3.246
|
NM_001252335
NM_032866
|
CGNL1
|
cingulin-like 1
|
|
chr3_-_18466759
|
3.032
|
NM_001195470
NM_002971
|
SATB1
|
SATB homeobox 1
|
|
chr3_-_18480203
|
2.980
|
NM_001131010
|
SATB1
|
SATB homeobox 1
|
|
chr17_+_68165660
|
2.883
|
NM_000891
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2
|
|
chr3_-_168864066
|
2.538
|
NM_001105077
NM_001163999
NM_005241
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr3_-_168865521
|
2.535
|
NM_001164000
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr3_-_168864325
|
2.533
|
NM_001105078
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr22_+_31518923
|
1.922
|
NM_001002837
|
INPP5J
|
inositol polyphosphate-5-phosphatase J
|
|
chr8_-_30891230
|
1.902
|
NM_001015508
|
PURG
|
purine-rich element binding protein G
|
|
chr1_+_61542928
|
1.782
|
NM_001145511
|
NFIA
|
nuclear factor I/A
|
|
chr1_+_61547625
|
1.760
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr3_-_169381417
|
1.613
|
NM_001205194
NM_004991
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr6_+_143999058
|
1.591
|
NM_001100164
NM_001100165
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr3_-_128212027
|
1.591
|
NM_032638
|
GATA2
|
GATA binding protein 2
|
|
chr9_-_14398981
|
1.585
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chr1_+_61547388
|
1.583
|
NM_001145512
|
NFIA
|
nuclear factor I/A
|
|
chr1_+_65730423
|
1.493
|
NM_014787
|
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6
|
|
chr3_-_128207366
|
1.490
|
NM_001145661
|
GATA2
|
GATA binding protein 2
|
|
chr9_-_14314036
|
1.442
|
NM_001190737
NM_005596
|
NFIB
|
nuclear factor I/B
|
|
chr6_+_143929316
|
1.430
|
NM_001100166
NM_014721
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr17_+_61086897
|
1.327
|
NM_025185
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2
|
|
chr12_-_105629790
|
1.268
|
NM_001251904
NM_018171
|
APPL2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2
|
|
chr17_-_66287256
|
1.229
|
NM_001174166
NM_004694
|
SLC16A6
|
solute carrier family 16, member 6 (monocarboxylic acid transporter 7)
|
|
chr3_+_150126705
|
1.170
|
NM_014779
|
TSC22D2
|
TSC22 domain family, member 2
|
|
chr7_+_155089483
|
1.123
|
NM_005542
NM_198336
NM_198337
|
INSIG1
|
insulin induced gene 1
|
|
chr6_+_107811272
|
1.087
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr19_-_14316980
|
1.069
|
NM_001008701
NM_014921
|
LPHN1
|
latrophilin 1
|
|
chr21_-_40033617
|
1.067
|
NM_001243432
NM_001136154
NM_004449
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr15_+_57210823
|
1.034
|
NM_003205
NM_207036
NM_207037
NM_207038
|
TCF12
|
transcription factor 12
|
|
chr21_-_39870303
|
1.007
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr12_+_65004191
|
1.005
|
NM_178169
|
RASSF3
|
Ras association (RalGDS/AF-6) domain family member 3
|
|
chr2_+_113033146
|
1.003
|
NM_198581
|
ZC3H6
|
zinc finger CCCH-type containing 6
|
|
chr7_-_143582363
|
0.989
|
NM_001206941
|
FAM115A
|
family with sequence similarity 115, member A
|
|
chr11_+_86748870
|
0.946
|
NM_001168724
NM_022918
|
TMEM135
|
transmembrane protein 135
|
|
chr11_-_130184459
|
0.940
|
NM_014155
|
ZBTB44
|
zinc finger and BTB domain containing 44
|
|
chr13_-_107187312
|
0.917
|
NM_004093
|
EFNB2
|
ephrin-B2
|
|
chr1_+_244216506
|
0.916
|
NM_006352
|
ZNF238
|
zinc finger protein 238
|
|
chr15_+_86220267
|
0.893
|
NM_144767
|
AKAP13
|
A kinase (PRKA) anchor protein 13
|
|
chr14_+_64680858
|
0.887
|
NM_182913
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2
|
|
chr11_+_9596233
|
0.879
|
NM_001143976
|
WEE1
|
WEE1 homolog (S. pombe)
|
|
chr10_-_21814610
|
0.870
|
NM_207371
|
C10orf140
|
chromosome 10 open reading frame 140
|
|
chr12_-_12419702
|
0.861
|
NM_002336
|
LRP6
|
low density lipoprotein receptor-related protein 6
|
|
chr6_+_42788793
|
0.860
|
NM_015349
|
KIAA0240
|
KIAA0240
|
|
chr9_-_125027080
|
0.858
|
NM_033117
|
RBM18
|
RNA binding motif protein 18
|
|
chr14_+_64682671
|
0.854
|
NM_182910
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2
|
|
chrX_-_63005212
|
0.841
|
NM_001173479
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chr15_+_57511616
|
0.825
|
NM_207040
|
TCF12
|
transcription factor 12
|
|
chr7_-_143599163
|
0.807
|
NM_014719
NM_001206938
|
FAM115A
|
family with sequence similarity 115, member A
|
|
chr1_-_179112178
|
0.798
|
NM_001136000
NM_001168239
NM_005158
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr1_+_244214552
|
0.773
|
NM_205768
|
ZNF238
|
zinc finger protein 238
|
|
chr3_-_114866090
|
0.750
|
NM_015642
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr11_+_9595183
|
0.732
|
NM_003390
|
WEE1
|
WEE1 homolog (S. pombe)
|
|
chr9_-_126030854
|
0.722
|
NM_001171137
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr19_-_47617008
|
0.720
|
NM_015168
|
ZC3H4
|
zinc finger CCCH-type containing 4
|
|
chr17_-_73975507
|
0.707
|
NM_001185039
NM_004035
NM_007292
|
ACOX1
|
acyl-CoA oxidase 1, palmitoyl
|
|
chr7_+_12726413
|
0.707
|
NM_001037164
NM_212460
|
ARL4A
|
ADP-ribosylation factor-like 4A
|
|
chr17_+_31254927
|
0.693
|
NM_001033504
NM_015544
|
TMEM98
|
transmembrane protein 98
|
|
chr8_-_81787015
|
0.691
|
NM_001033723
|
ZNF704
|
zinc finger protein 704
|
|
chr9_-_126030792
|
0.691
|
NM_018387
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr7_+_12726910
|
0.683
|
NM_001195396
NM_005738
|
ARL4A
|
ADP-ribosylation factor-like 4A
|
|
chr2_+_61404549
|
0.681
|
NM_152392
|
AHSA2
|
AHA1, activator of heat shock 90kDa protein ATPase homolog 2 (yeast)
|
|
chr4_-_152147520
|
0.679
|
NM_001009555
NM_001128923
|
SH3D19
|
SH3 domain containing 19
|
|
chr19_+_14016955
|
0.668
|
NM_017721
|
CC2D1A
|
coiled-coil and C2 domain containing 1A
|
|
chrX_-_62975005
|
0.666
|
NM_001173480
NM_015185
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chr5_+_172261220
|
0.665
|
NM_001031711
|
ERGIC1
|
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1
|
|
chr9_+_36036900
|
0.653
|
NM_021111
|
RECK
|
reversion-inducing-cysteine-rich protein with kazal motifs
|
|
chr12_+_27397077
|
0.652
|
NM_015000
|
STK38L
|
serine/threonine kinase 38 like
|
|
chr2_-_20251788
|
0.648
|
NM_014713
|
LAPTM4A
|
lysosomal protein transmembrane 4 alpha
|
|
chr6_-_16761600
|
0.647
|
NM_000332
NM_001128164
|
ATXN1
|
ataxin 1
|
|
chr2_+_42275017
|
0.614
|
NM_138370
|
PKDCC
|
protein kinase domain containing, cytoplasmic homolog (mouse)
|
|
chr12_-_109125289
|
0.609
|
NM_014325
|
CORO1C
|
coronin, actin binding protein, 1C
|
|
chr7_-_129592794
|
0.602
|
NM_003344
NM_182697
|
UBE2H
|
ubiquitin-conjugating enzyme E2H
|
|
chr11_-_124806302
|
0.599
|
NM_152722
|
HEPACAM
|
hepatic and glial cell adhesion molecule
|
|
chr2_-_122406994
|
0.588
|
NM_001142273
NM_001142274
NM_001207051
NM_015282
|
CLASP1
|
cytoplasmic linker associated protein 1
|
|
chr12_-_42538432
|
0.581
|
NM_001099650
NM_173601
|
GXYLT1
|
glucoside xylosyltransferase 1
|
|
chr1_-_179198814
|
0.577
|
NM_001136001
NM_001168236
NM_001168237
NM_001168238
NM_007314
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr22_-_37882214
|
0.576
|
NM_001166343
NM_002405
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr10_-_35103894
|
0.574
|
NM_001184785
NM_001184786
NM_001184787
NM_001184788
NM_001184789
NM_001184790
NM_001184791
NM_001184792
NM_001184793
NM_001184794
NM_019619
|
PARD3
|
par-3 partitioning defective 3 homolog (C. elegans)
|
|
chr1_+_28764660
|
0.568
|
NM_023923
|
PHACTR4
|
phosphatase and actin regulator 4
|
|
chr3_-_125094003
|
0.555
|
NM_021964
|
ZNF148
|
zinc finger protein 148
|
|
chr12_+_49209300
|
0.550
|
NM_001206916
|
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit
|
|
chr4_+_30721865
|
0.538
|
NM_032456
NM_001173523
NM_002589
NM_032457
|
PCDH7
|
protocadherin 7
|
|
chr7_-_129592711
|
0.537
|
NM_001202498
|
UBE2H
|
ubiquitin-conjugating enzyme E2H
|
|
chr11_-_86666211
|
0.527
|
NM_012193
|
FZD4
|
frizzled family receptor 4
|
|
chr2_-_61245363
|
0.514
|
NM_144709
|
PUS10
|
pseudouridylate synthase 10
|
|
chrX_-_135338640
|
0.513
|
NM_001173516
|
MAP7D3
|
MAP7 domain containing 3
|
|
chr1_+_28696056
|
0.504
|
NM_001048183
|
PHACTR4
|
phosphatase and actin regulator 4
|
|
chr5_-_14871555
|
0.501
|
NM_054027
|
ANKH
|
ankylosis, progressive homolog (mouse)
|
|
chr1_-_229478687
|
0.491
|
NM_145257
|
C1orf96
|
chromosome 1 open reading frame 96
|
|
chr9_+_37800779
|
0.481
|
NM_024345
|
DCAF10
|
DDB1 and CUL4 associated factor 10
|
|
chr10_-_75634219
|
0.469
|
NM_001204492
NM_001222
NM_172169
NM_172170
NM_172171
NM_172173
|
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma
|
|
chr6_+_151646634
|
0.468
|
NM_144497
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr20_-_31071187
|
0.467
|
NM_080616
|
C20orf112
|
chromosome 20 open reading frame 112
|
|
chr3_-_114343052
|
0.464
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr2_-_68547018
|
0.454
|
NM_001111101
NM_015463
|
CNRIP1
|
cannabinoid receptor interacting protein 1
|
|
chr1_-_115632047
|
0.451
|
NM_005725
|
TSPAN2
|
tetraspanin 2
|
|
chr1_+_201708940
|
0.449
|
NM_001167738
|
NAV1
|
neuron navigator 1
|
|
chr4_-_152096655
|
0.443
|
NM_001128924
|
SH3D19
|
SH3 domain containing 19
|
|
chr4_+_141445311
|
0.440
|
NM_153702
|
ELMOD2
|
ELMO/CED-12 domain containing 2
|
|
chr6_+_151561094
|
0.439
|
NM_005100
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr1_-_225840660
|
0.430
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr1_-_154943001
|
0.428
|
NM_001130040
NM_183001
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr6_-_166582129
|
0.425
|
NM_003181
|
T
|
T, brachyury homolog (mouse)
|
|
chr5_+_118407067
|
0.416
|
NM_005509
|
DMXL1
|
Dmx-like 1
|
|
chr4_-_77134987
|
0.413
|
NM_001204255
NM_005506
|
SCARB2
|
scavenger receptor class B, member 2
|
|
chr3_+_169940214
|
0.406
|
NM_002740
|
PRKCI
|
protein kinase C, iota
|
|
chr4_+_57775064
|
0.406
|
NM_001193508
|
REST
|
RE1-silencing transcription factor
|
|
chr12_+_49208214
|
0.404
|
NM_001206917
|
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit
|
|
chr19_-_2051222
|
0.404
|
NM_017572
NM_199054
|
MKNK2
|
MAP kinase interacting serine/threonine kinase 2
|
|
chr16_+_69139466
|
0.387
|
NM_001199280
|
HAS3
|
hyaluronan synthase 3
|
|
chr2_-_171923482
|
0.385
|
NM_001136555
|
TLK1
|
tousled-like kinase 1
|
|
chr15_+_52043738
|
0.380
|
NM_001142885
NM_014548
|
TMOD2
|
tropomodulin 2 (neuronal)
|
|
chr13_+_108870713
|
0.377
|
NM_032859
|
ABHD13
|
abhydrolase domain containing 13
|
|
chr5_+_78531848
|
0.377
|
NM_152405
|
JMY
|
junction mediating and regulatory protein, p53 cofactor
|
|
chr7_+_17338182
|
0.376
|
NM_001621
|
AHR
|
aryl hydrocarbon receptor
|
|
chr2_-_172087823
|
0.375
|
NM_001136554
|
TLK1
|
tousled-like kinase 1
|
|
chr16_+_69141442
|
0.374
|
NM_005329
|
HAS3
|
hyaluronan synthase 3
|
|
chr3_-_114343791
|
0.369
|
NM_001164346
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chrX_+_129139163
|
0.365
|
NM_021946
|
BCORL1
|
BCL6 corepressor-like 1
|
|
chr15_+_69452799
|
0.362
|
NM_015554
|
GLCE
|
glucuronic acid epimerase
|
|
chr3_-_128206704
|
0.356
|
NM_001145662
|
GATA2
|
GATA binding protein 2
|
|
chr19_-_17414083
|
0.353
|
NM_024527
|
ABHD8
|
abhydrolase domain containing 8
|
|
chr3_-_114478117
|
0.350
|
NM_001164344
NM_001164345
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr14_+_60712469
|
0.349
|
NM_177952
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A
|
|
chr4_+_57774035
|
0.347
|
NM_005612
|
REST
|
RE1-silencing transcription factor
|
|
chr7_+_21467572
|
0.342
|
NM_003112
|
SP4
|
Sp4 transcription factor
|
|
chr19_-_11308184
|
0.335
|
NM_001136191
|
KANK2
|
KN motif and ankyrin repeat domains 2
|
|
chr15_-_23932410
|
0.334
|
NM_002487
|
NDN
|
necdin homolog (mouse)
|
|
chr2_-_172017358
|
0.331
|
NM_012290
|
TLK1
|
tousled-like kinase 1
|
|
chr6_-_119031230
|
0.330
|
NM_001178035
|
CEP85L
|
centrosomal protein 85kDa-like
|
|
chr19_-_11305186
|
0.328
|
NM_015493
|
KANK2
|
KN motif and ankyrin repeat domains 2
|
|
chr4_-_122744978
|
0.328
|
NM_001237
|
CCNA2
|
cyclin A2
|
|
chr12_+_72233486
|
0.327
|
NM_001146213
NM_001146214
NM_022771
|
TBC1D15
|
TBC1 domain family, member 15
|
|
chr9_-_123555654
|
0.321
|
NM_012164
|
FBXW2
|
F-box and WD repeat domain containing 2
|
|
chr20_-_50418946
|
0.316
|
NM_020436
|
SALL4
|
sal-like 4 (Drosophila)
|
|
chr8_+_17013799
|
0.313
|
NM_016353
|
ZDHHC2
|
zinc finger, DHHC-type containing 2
|
|
chr1_+_150039341
|
0.309
|
NM_007259
|
VPS45
|
vacuolar protein sorting 45 homolog (S. cerevisiae)
|
|
chr11_+_120207617
|
0.304
|
NM_001198665
NM_015313
|
ARHGEF12
|
Rho guanine nucleotide exchange factor (GEF) 12
|
|
chr12_+_49212253
|
0.302
|
NM_000725
NM_001206915
|
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit
|
|
chr1_+_180601121
|
0.298
|
NM_001135669
NM_004736
|
XPR1
|
xenotropic and polytropic retrovirus receptor 1
|
|
chr19_+_5914192
|
0.296
|
NM_004058
NM_080590
|
CAPS
|
calcyphosine
|
|
chrX_+_107334896
|
0.294
|
NM_052936
NM_178270
|
ATG4A
|
ATG4 autophagy related 4 homolog A (S. cerevisiae)
|
|
chr7_-_138665853
|
0.294
|
NM_001164665
NM_020910
|
KIAA1549
|
KIAA1549
|
|
chr14_-_55878537
|
0.292
|
NM_014924
|
ATG14
|
ATG14 autophagy related 14 homolog (S. cerevisiae)
|
|
chr16_+_58534045
|
0.290
|
NM_001242833
NM_001242834
NM_001242835
NM_001242836
|
NDRG4
|
NDRG family member 4
|
|
chr6_-_151773168
|
0.285
|
NM_017909
|
RMND1
|
required for meiotic nuclear division 1 homolog (S. cerevisiae)
|
|
chr18_+_905296
|
0.284
|
NM_001117
|
ADCYAP1
|
adenylate cyclase activating polypeptide 1 (pituitary)
|
|
chr2_+_196521851
|
0.283
|
NM_020342
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10
|
|
chr3_+_14989076
|
0.276
|
NM_003298
|
NR2C2
|
nuclear receptor subfamily 2, group C, member 2
|
|
chr14_+_102228134
|
0.269
|
NM_001161725
NM_001161726
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr17_-_18266691
|
0.259
|
NM_004169
NM_148918
|
SHMT1
|
serine hydroxymethyltransferase 1 (soluble)
|
|
chr14_+_71996041
|
0.258
|
NM_015556
|
SIPA1L1
|
signal-induced proliferation-associated 1 like 1
|
|
chr6_+_152128453
|
0.256
|
NM_001122740
|
ESR1
|
estrogen receptor 1
|
|
chr6_+_17282931
|
0.255
|
NM_153020
|
RBM24
|
RNA binding motif protein 24
|
|
chr3_-_38691118
|
0.250
|
NM_000335
NM_001099404
NM_001099405
NM_198056
NM_001160160
NM_001160161
|
SCN5A
|
sodium channel, voltage-gated, type V, alpha subunit
|
|
chr12_-_49449106
|
0.249
|
NM_003482
|
MLL2
|
myeloid/lymphoid or mixed-lineage leukemia 2
|
|
chrX_-_54209639
|
0.245
|
NM_017848
NM_198456
|
FAM120C
|
family with sequence similarity 120C
|
|
chr3_-_114102488
|
0.241
|
NM_001164342
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr20_+_4666796
|
0.239
|
NM_000311
NM_001080121
NM_001080122
NM_183079
NM_001080123
|
PRNP
|
prion protein
|
|
chr11_-_33795892
|
0.234
|
NM_012175
NM_033406
|
FBXO3
|
F-box protein 3
|
|
chr10_+_73975752
|
0.234
|
NM_001242546
NM_001242547
NM_001242548
NM_173473
|
ANAPC16
|
anaphase promoting complex subunit 16
|
|
chr15_+_91411884
|
0.233
|
NM_002569
|
FURIN
|
furin (paired basic amino acid cleaving enzyme)
|
|
chr1_-_42800902
|
0.223
|
NM_014947
|
FOXJ3
|
forkhead box J3
|
|
chr11_-_84028379
|
0.221
|
NM_001206769
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr1_-_42800883
|
0.220
|
NM_001198852
|
FOXJ3
|
forkhead box J3
|
|
chr5_+_65440004
|
0.219
|
NM_001077199
|
SREK1
|
splicing regulatory glutamine/lysine-rich protein 1
|
|
chr22_-_21213099
|
0.213
|
NM_058004
|
PI4KA
|
phosphatidylinositol 4-kinase, catalytic, alpha
|
|
chr20_-_48732479
|
0.213
|
NM_022442
NM_021988
NM_199144
|
UBE2V1
|
ubiquitin-conjugating enzyme E2 variant 1
|
|
chr6_+_72922513
|
0.206
|
NM_001168407
NM_001168408
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr20_-_48729664
|
0.206
|
NM_001032288
|
UBE2V1
|
ubiquitin-conjugating enzyme E2 variant 1
|
|
chr12_+_58087737
|
0.205
|
NM_001017956
NM_001017957
NM_001017958
NM_006812
|
OS9
|
osteosarcoma amplified 9, endoplasmic reticulum lectin
|
|
chr16_+_57126439
|
0.204
|
NM_152727
|
CPNE2
|
copine II
|
|
chr22_-_21088954
|
0.202
|
NM_002650
|
PI4KA
|
phosphatidylinositol 4-kinase, catalytic, alpha
|
|
chr6_+_139094656
|
0.202
|
NM_015439
|
CCDC28A
|
coiled-coil domain containing 28A
|
|
chr16_+_58498725
|
0.194
|
NM_022910
|
NDRG4
|
NDRG family member 4
|
|
chr5_+_65440662
|
0.191
|
NM_139168
|
SREK1
|
splicing regulatory glutamine/lysine-rich protein 1
|
|
chr7_+_30323922
|
0.190
|
NM_147128
|
ZNRF2
|
zinc and ring finger 2
|
|
chr1_+_33283042
|
0.190
|
NM_001017406
NM_022753
|
S100PBP
|
S100P binding protein
|
|
chr17_-_29151724
|
0.186
|
NM_015986
|
CRLF3
|
cytokine receptor-like factor 3
|
|
chr19_+_42829760
|
0.185
|
NM_001410
|
MEGF8
|
multiple EGF-like-domains 8
|
|
chr9_+_102584136
|
0.184
|
NM_006981
NM_173199
NM_173200
|
NR4A3
|
nuclear receptor subfamily 4, group A, member 3
|
|
chr6_+_150070818
|
0.183
|
NM_001252049
NM_001252050
NM_001252051
NM_001252052
NM_001252053
NM_005389
|
PCMT1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase
|
|
chr7_-_5463176
|
0.180
|
NM_001080495
|
TNRC18
|
trinucleotide repeat containing 18
|
|
chr3_-_114790221
|
0.179
|
NM_001164343
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr7_+_106809405
|
0.178
|
NM_001244262
NM_012257
|
HBP1
|
HMG-box transcription factor 1
|
|
chr16_+_58497993
|
0.175
|
NM_001130487
|
NDRG4
|
NDRG family member 4
|
|
chr6_+_167412791
|
0.171
|
NM_007045
NM_194429
|
FGFR1OP
|
FGFR1 oncogene partner
|
|
chr1_+_29562947
|
0.169
|
NM_001195001
NM_005704
NM_133177
NM_133178
|
PTPRU
|
protein tyrosine phosphatase, receptor type, U
|
|
chr2_+_196521470
|
0.169
|
NM_001127257
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10
|
|
chr1_+_201617449
|
0.168
|
NM_020443
|
NAV1
|
neuron navigator 1
|
|
chr19_-_3700466
|
0.167
|
NM_001195733
NM_012398
|
PIP5K1C
|
phosphatidylinositol-4-phosphate 5-kinase, type I, gamma
|
|
chr5_-_74807805
|
0.166
|
NM_001130105
|
COL4A3BP
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein
|
|
chr8_-_55014367
|
0.165
|
NM_006330
|
LYPLA1
|
lysophospholipase I
|