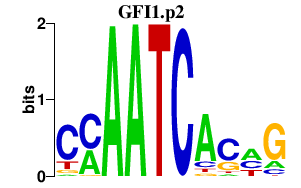
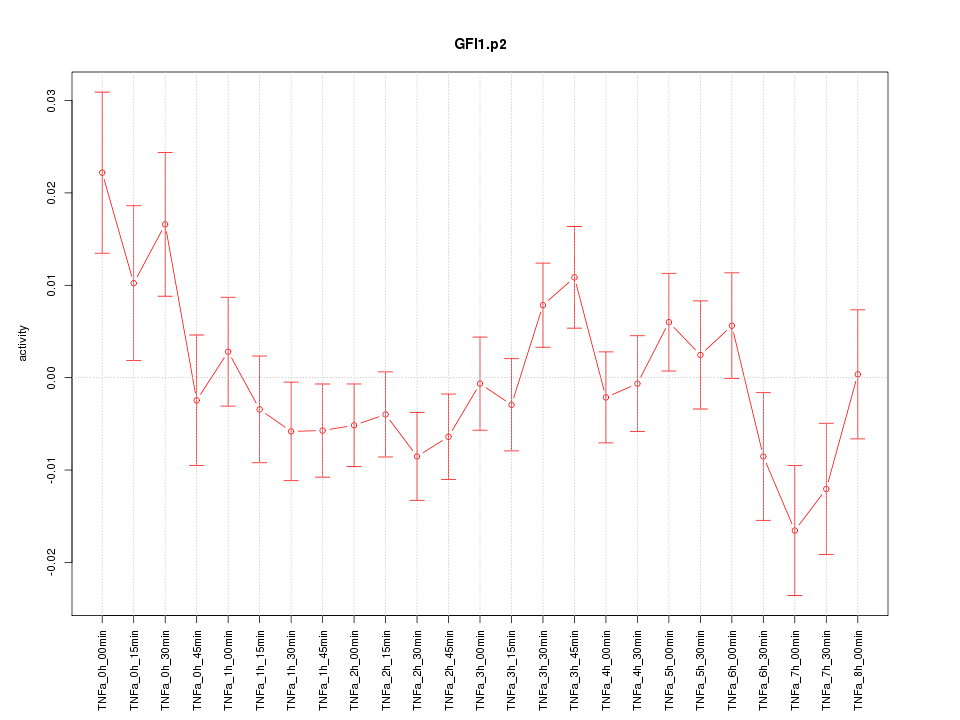
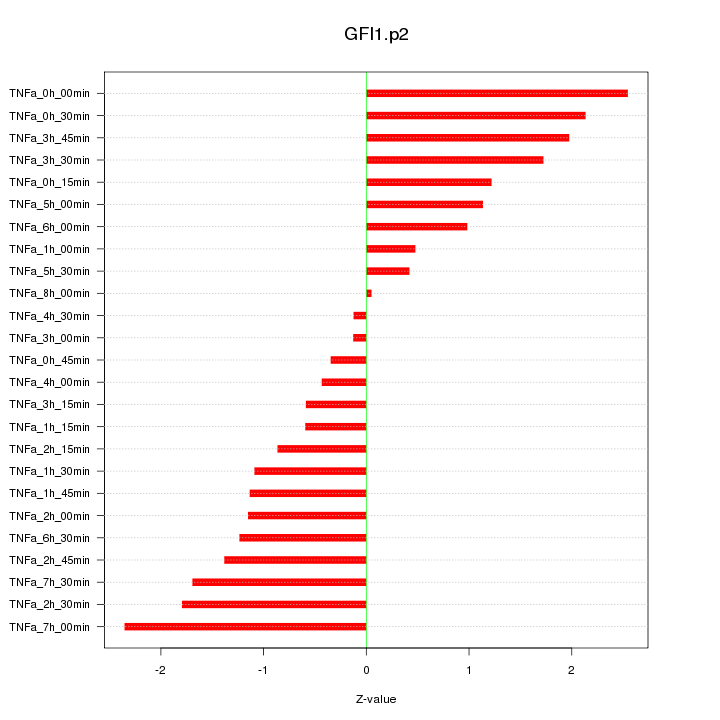
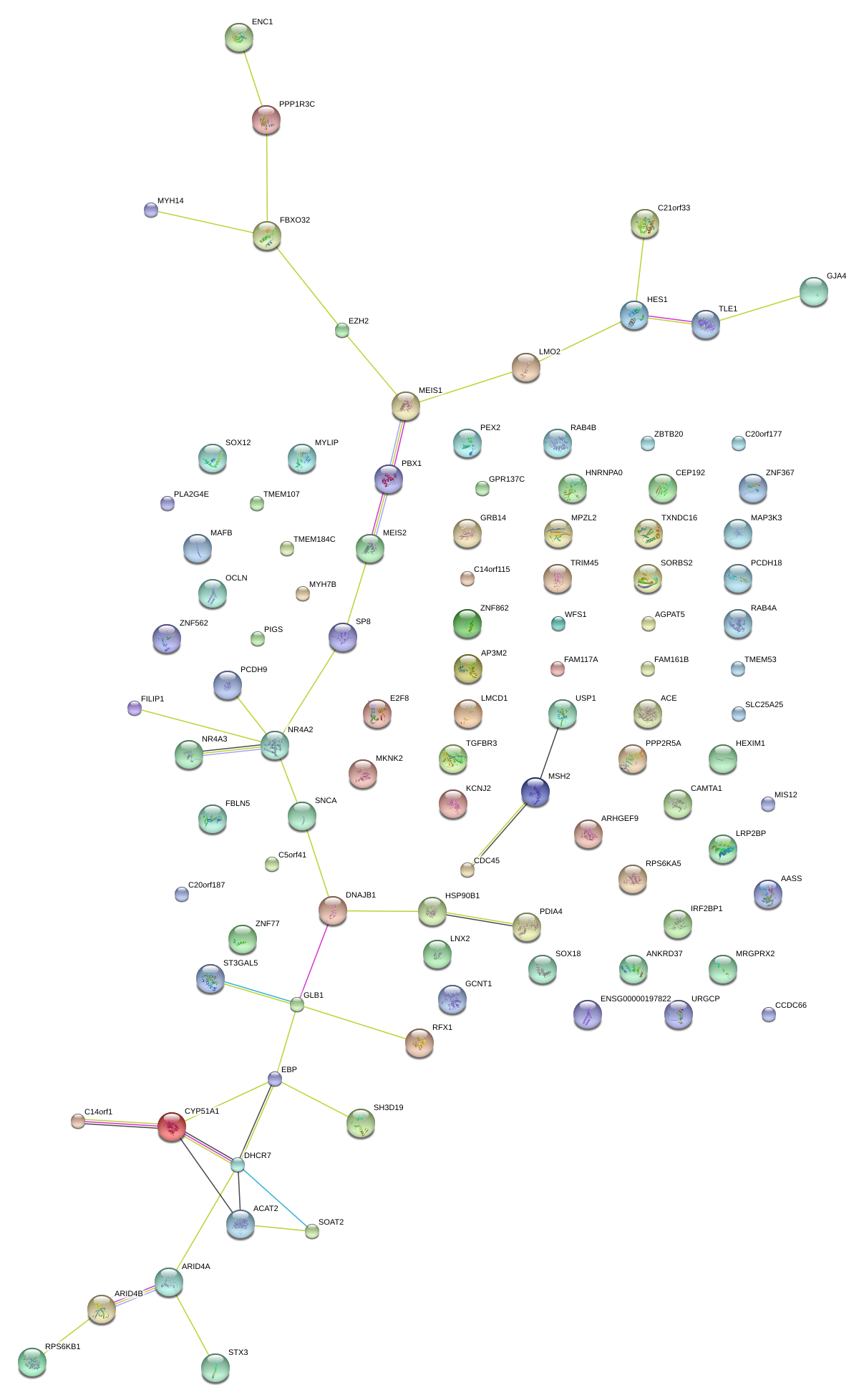
|
chr9_+_79093256
|
2.567
|
NM_001097635
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chr14_+_58765209
|
2.421
|
NM_002892
NM_023000
NM_023001
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like)
|
|
chr20_-_62680880
|
2.331
|
NM_018419
|
SOX18
|
SRY (sex determining region Y)-box 18
|
|
chr20_+_58508818
|
2.295
|
NM_001190826
|
C20orf177
|
chromosome 20 open reading frame 177
|
|
chr1_-_45140117
|
2.239
|
|
TMEM53
|
transmembrane protein 53
|
|
chr17_-_47841517
|
2.180
|
NM_030802
|
FAM117A
|
family with sequence similarity 117, member A
|
|
chr10_-_93392744
|
2.172
|
|
PPP1R3C
|
protein phosphatase 1, regulatory subunit 3C
|
|
chr10_-_93392857
|
2.163
|
NM_005398
|
PPP1R3C
|
protein phosphatase 1, regulatory subunit 3C
|
|
chr7_-_148725500
|
1.898
|
|
PDIA4
|
protein disulfide isomerase family A, member 4
|
|
chr17_-_47841468
|
1.882
|
|
FAM117A
|
family with sequence similarity 117, member A
|
|
chr4_-_186300151
|
1.819
|
NM_018409
|
LRP2BP
|
LRP2 binding protein
|
|
chr14_+_58765275
|
1.785
|
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like)
|
|
chr14_-_91526912
|
1.754
|
NM_004755
NM_182398
|
RPS6KA5
|
ribosomal protein S6 kinase, 90kDa, polypeptide 5
|
|
chr14_-_74416711
|
1.542
|
|
FAM161B
|
family with sequence similarity 161, member B
|
|
chr4_-_138453628
|
1.518
|
NM_019035
|
PCDH18
|
protocadherin 18
|
|
chr1_-_45140062
|
1.479
|
NM_024587
|
TMEM53
|
transmembrane protein 53
|
|
chr17_+_68165660
|
1.469
|
NM_000891
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2
|
|
chr1_+_164528586
|
1.459
|
NM_001204961
NM_001204963
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr19_-_14640048
|
1.452
|
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1
|
|
chr4_+_6271558
|
1.442
|
NM_001145853
NM_006005
|
WFS1
|
Wolfram syndrome 1 (wolframin)
|
|
chr4_+_6271588
|
1.409
|
|
WFS1
|
Wolfram syndrome 1 (wolframin)
|
|
chr4_+_6271605
|
1.405
|
|
WFS1
|
Wolfram syndrome 1 (wolframin)
|
|
chr2_-_165424993
|
1.399
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr14_-_58764805
|
1.353
|
|
FLJ31306
|
uncharacterized LOC379025
|
|
chr14_-_53019036
|
1.315
|
NM_001160047
NM_020784
|
TXNDC16
|
thioredoxin domain containing 16
|
|
chr7_-_148725731
|
1.305
|
|
PDIA4
|
protein disulfide isomerase family A, member 4
|
|
chr7_-_121784236
|
1.289
|
NM_005763
|
AASS
|
aminoadipate-semialdehyde synthase
|
|
chr7_-_148725764
|
1.288
|
NM_004911
|
PDIA4
|
protein disulfide isomerase family A, member 4
|
|
chr19_-_2042022
|
1.248
|
|
MKNK2
|
MAP kinase interacting serine/threonine kinase 2
|
|
chr1_+_212458867
|
1.223
|
NM_006243
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha
|
|
chr3_+_8543508
|
1.204
|
NM_014583
|
LMCD1
|
LIM and cysteine-rich domains 1
|
|
chr7_+_149535478
|
1.170
|
|
ZNF862
|
zinc finger protein 862
|
|
chr7_-_148581359
|
1.169
|
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr15_-_23378217
|
1.131
|
|
HERC2P2
|
hect domain and RLD 2 pseudogene 2
|
|
chr1_-_92351476
|
1.111
|
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr6_+_160183062
|
1.103
|
|
ACAT2
|
acetyl-CoA acetyltransferase 2
|
|
chr6_+_160182988
|
1.097
|
NM_005891
|
ACAT2
|
acetyl-CoA acetyltransferase 2
|
|
chr6_+_160183510
|
1.097
|
|
ACAT2
|
acetyl-CoA acetyltransferase 2
|
|
chr3_-_114866090
|
1.097
|
NM_015642
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr7_-_148581297
|
1.093
|
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr5_+_68800003
|
1.083
|
NM_001205255
|
OCLN
|
occludin
|
|
chr11_-_19262424
|
1.069
|
NM_024680
|
E2F8
|
E2F transcription factor 8
|
|
chr19_-_14116875
|
1.068
|
|
RFX1
|
regulatory factor X, 1 (influences HLA class II expression)
|
|
chr19_-_2944894
|
1.060
|
NM_021217
|
ZNF77
|
zinc finger protein 77
|
|
chr4_-_138453564
|
1.049
|
|
PCDH18
|
protocadherin 18
|
|
chr3_+_56591182
|
1.046
|
NM_001012506
NM_001141947
|
CCDC66
|
coiled-coil domain containing 66
|
|
chr17_+_61699786
|
1.044
|
NM_002401
NM_203351
|
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3
|
|
chr4_-_186733372
|
1.034
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr6_-_76203256
|
1.027
|
NM_015687
|
FILIP1
|
filamin A interacting protein 1
|
|
chr2_+_47630291
|
1.020
|
|
MSH2
|
mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli)
|
|
chr3_+_183165402
|
1.016
|
|
LOC100505687
|
uncharacterized LOC100505687
|
|
chr9_-_84304378
|
1.009
|
|
TLE1
|
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila)
|
|
chr22_+_19467413
|
1.007
|
NM_001178010
NM_001178011
NM_003504
|
CDC45
|
cell division cycle 45 homolog (S. cerevisiae)
|
|
chr2_+_47630190
|
0.998
|
|
MSH2
|
mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli)
|
|
chr2_+_47630246
|
0.997
|
NM_000251
|
MSH2
|
mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli)
|
|
chr19_-_2041862
|
0.994
|
|
MKNK2
|
MAP kinase interacting serine/threonine kinase 2
|
|
chrX_+_48380163
|
0.973
|
NM_006579
|
EBP
|
emopamil binding protein (sterol isomerase)
|
|
chr1_+_62902325
|
0.963
|
NM_003368
|
USP1
|
ubiquitin specific peptidase 1
|
|
chr2_-_86116016
|
0.959
|
NM_003896
|
ST3GAL5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5
|
|
chr1_+_6845381
|
0.957
|
NM_001195563
NM_001242701
NM_015215
|
CAMTA1
|
calmodulin binding transcription activator 1
|
|
chr11_-_118135197
|
0.950
|
NM_005797
NM_144765
|
MPZL2
|
myelin protein zero-like 2
|
|
chr14_-_76127158
|
0.946
|
|
C14orf1
|
chromosome 14 open reading frame 1
|
|
chr22_+_19467467
|
0.939
|
|
CDC45
|
cell division cycle 45 homolog (S. cerevisiae)
|
|
chr5_-_137090049
|
0.937
|
|
|
|
|
chr5_-_137090033
|
0.916
|
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0
|
|
chr19_-_14629160
|
0.904
|
NM_006145
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1
|
|
chr17_+_5389864
|
0.903
|
|
MIS12
|
MIS12, MIND kinetochore complex component, homolog (S. pombe)
|
|
chr1_+_35258598
|
0.885
|
NM_002060
|
GJA4
|
gap junction protein, alpha 4, 37kDa
|
|
chr11_-_19082227
|
0.882
|
NM_054030
|
MRGPRX2
|
MAS-related GPR, member X2
|
|
chr17_+_43225834
|
0.879
|
|
HEXIM1
|
hexamethylene bis-acetamide inducible 1
|
|
chr7_-_20826503
|
0.879
|
NM_182700
NM_198956
|
SP8
|
Sp8 transcription factor
|
|
chr9_+_130830447
|
0.873
|
NM_001006641
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr20_-_39317875
|
0.871
|
NM_005461
|
MAFB
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian)
|
|
chr14_-_92414004
|
0.867
|
NM_006329
|
FBLN5
|
fibulin 5
|
|
chr2_+_66662692
|
0.866
|
|
MEIS1
|
Meis homeobox 1
|
|
chr17_+_61554420
|
0.860
|
NM_000789
|
ACE
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1
|
|
chr6_+_16129285
|
0.857
|
NM_013262
|
MYLIP
|
myosin regulatory light chain interacting protein
|
|
chr14_-_76127526
|
0.840
|
NM_007176
|
C14orf1
|
chromosome 14 open reading frame 1
|
|
chr11_-_33890931
|
0.835
|
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr13_-_67804432
|
0.831
|
NM_020403
NM_203487
|
PCDH9
|
protocadherin 9
|
|
chr11_-_71159371
|
0.827
|
|
DHCR7
|
7-dehydrocholesterol reductase
|
|
chr9_+_127420690
|
0.820
|
|
MIR181A2HG
|
MIR181A2 host gene (non-protein coding)
|
|
chr18_+_12991329
|
0.818
|
NM_032142
|
CEP192
|
centrosomal protein 192kDa
|
|
chr19_-_14629227
|
0.818
|
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1
|
|
chr2_-_157189040
|
0.817
|
NM_006186
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chr12_+_104324157
|
0.817
|
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1
|
|
chr18_-_22930826
|
0.813
|
|
|
|
|
chr9_-_99180661
|
0.812
|
NM_153695
|
ZNF367
|
zinc finger protein 367
|
|
chr19_-_46389307
|
0.808
|
NM_015649
|
IRF2BP1
|
interferon regulatory factor 2 binding protein 1
|
|
chr1_-_117664349
|
0.804
|
NM_001145635
NM_025188
|
TRIM45
|
tripartite motif containing 45
|
|
chr13_-_28194677
|
0.803
|
NM_153371
|
LNX2
|
ligand of numb-protein X 2
|
|
chr14_+_53019865
|
0.796
|
NM_001099652
|
GPR137C
|
G protein-coupled receptor 137C
|
|
chr19_+_50706868
|
0.794
|
NM_001077186
NM_001145809
NM_024729
|
MYH14
|
myosin, heavy chain 14, non-muscle
|
|
chr17_+_61699715
|
0.794
|
|
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3
|
|
chr4_+_148538564
|
0.794
|
|
TMEM184C
|
transmembrane protein 184C
|
|
chr11_-_71159442
|
0.786
|
NM_001163817
NM_001360
|
DHCR7
|
7-dehydrocholesterol reductase
|
|
chr7_-_148581379
|
0.777
|
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr17_-_26898495
|
0.772
|
|
PIGS
|
phosphatidylinositol glycan anchor biosynthesis, class S
|
|
chr8_+_42010463
|
0.770
|
NM_001134296
NM_006803
|
AP3M2
|
adaptor-related protein complex 3, mu 2 subunit
|
|
chr5_-_137089990
|
0.770
|
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0
|
|
chr20_+_306205
|
0.766
|
NM_006943
|
SOX12
|
SRY (sex determining region Y)-box 12
|
|
chr4_-_90759446
|
0.766
|
NM_001146055
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor)
|
|
chr3_+_193853930
|
0.764
|
NM_005524
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr19_+_41284068
|
0.760
|
NM_016154
|
RAB4B
|
RAB4B, member RAS oncogene family
|
|
chr17_+_5389715
|
0.758
|
|
MIS12
|
MIS12, MIND kinetochore complex component, homolog (S. pombe)
|
|
chr3_+_193853937
|
0.757
|
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr11_+_59522531
|
0.756
|
NM_001178040
NM_004177
|
STX3
|
syntaxin 3
|
|
chr17_+_43225921
|
0.755
|
|
HEXIM1
|
hexamethylene bis-acetamide inducible 1
|
|
chr7_-_91763828
|
0.755
|
NM_000786
|
CYP51A1
|
cytochrome P450, family 51, subfamily A, polypeptide 1
|
|
chrX_-_62974942
|
0.755
|
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chr5_-_73937248
|
0.754
|
NM_003633
|
ENC1
|
ectodermal-neural cortex 1 (with BTB-like domain)
|
|
chr7_-_91763677
|
0.748
|
|
CYP51A1
|
cytochrome P450, family 51, subfamily A, polypeptide 1
|
|
chr13_-_28194538
|
0.747
|
|
LNX2
|
ligand of numb-protein X 2
|
|
chr8_-_77912296
|
0.747
|
NM_000318
NM_001079867
|
PEX2
|
peroxisomal biogenesis factor 2
|
|
chr11_-_19263144
|
0.745
|
|
E2F8
|
E2F transcription factor 8
|
|
chr8_+_6565870
|
0.743
|
NM_018361
|
AGPAT5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon)
|
|
chr7_-_91763775
|
0.739
|
|
CYP51A1
|
cytochrome P450, family 51, subfamily A, polypeptide 1
|
|
chr7_-_91764036
|
0.735
|
NM_001146152
|
CYP51A1
|
cytochrome P450, family 51, subfamily A, polypeptide 1
|
|
chr5_+_172483285
|
0.733
|
NM_001168393
NM_001168394
NM_153607
|
C5orf41
|
chromosome 5 open reading frame 41
|
|
chr6_+_16129321
|
0.731
|
|
MYLIP
|
myosin regulatory light chain interacting protein
|
|
chr2_-_73460326
|
0.729
|
NM_032319
|
PRADC1
|
protease-associated domain containing 1
|
|
chr8_-_124544376
|
0.727
|
NM_148177
|
FBXO32
|
F-box protein 32
|
|
chr11_-_4629371
|
0.724
|
NM_018073
|
TRIM68
|
tripartite motif containing 68
|
|
chrX_-_52987610
|
0.724
|
NM_001242491
|
FAM156A
FAM156B
|
family with sequence similarity 156, member A
family with sequence similarity 156, member B
|
|
chr8_+_6566153
|
0.721
|
|
AGPAT5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon)
|
|
chr9_-_75567915
|
0.720
|
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|
|
chr4_+_41614911
|
0.719
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr1_-_92351613
|
0.718
|
NM_001195683
NM_003243
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr1_+_63249771
|
0.717
|
NM_032852
NM_178221
|
ATG4C
|
ATG4 autophagy related 4 homolog C (S. cerevisiae)
|
|
chr9_+_130830595
|
0.708
|
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr3_-_148804122
|
0.702
|
NM_003071
NM_139048
|
HLTF
|
helicase-like transcription factor
|
|
chr19_-_14247267
|
0.700
|
|
ASF1B
|
ASF1 anti-silencing function 1 homolog B (S. cerevisiae)
|
|
chr12_+_104324192
|
0.699
|
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1
|
|
chr5_-_100238870
|
0.697
|
NM_175052
NM_005668
|
ST8SIA4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4
|
|
chr12_+_104324186
|
0.696
|
NM_003299
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1
|
|
chrX_+_52926358
|
0.695
|
|
FAM156B
|
family with sequence similarity 156, member B
|
|
chr4_+_148538508
|
0.694
|
NM_018241
|
TMEM184C
|
transmembrane protein 184C
|
|
chrX_+_52926317
|
0.693
|
NM_001242491
|
FAM156A
|
family with sequence similarity 156, member A
|
|
chrX_+_52926334
|
0.690
|
|
FAM156A
FAM156B
|
family with sequence similarity 156, member A
family with sequence similarity 156, member B
|
|
chr7_+_149535508
|
0.689
|
NM_001099220
|
ZNF862
|
zinc finger protein 862
|
|
chr16_+_53164784
|
0.685
|
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr5_-_43313492
|
0.679
|
|
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble)
|
|
chr11_-_2906960
|
0.674
|
NM_000076
NM_001122630
NM_001122631
|
CDKN1C
|
cyclin-dependent kinase inhibitor 1C (p57, Kip2)
|
|
chr17_+_4981754
|
0.671
|
|
ZFP3
|
zinc finger protein 3 homolog (mouse)
|
|
chr8_-_103667882
|
0.671
|
NM_005655
|
KLF10
|
Kruppel-like factor 10
|
|
chr7_+_141490016
|
0.670
|
NM_018980
|
TAS2R5
|
taste receptor, type 2, member 5
|
|
chr12_+_104324214
|
0.670
|
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1
|
|
chr12_-_122240827
|
0.665
|
|
|
|
|
chr14_+_100150596
|
0.659
|
NM_006668
|
CYP46A1
|
cytochrome P450, family 46, subfamily A, polypeptide 1
|
|
chrX_-_62974987
|
0.658
|
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chr7_-_148581377
|
0.658
|
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr10_+_102133599
|
0.654
|
|
LINC00263
|
long intergenic non-protein coding RNA 263
|
|
chr2_+_191745791
|
0.652
|
|
GLS
|
glutaminase
|
|
chr3_+_20081523
|
0.647
|
NM_003884
|
KAT2B
|
K(lysine) acetyltransferase 2B
|
|
chr15_-_23378167
|
0.645
|
|
HERC2P2
|
hect domain and RLD 2 pseudogene 2
|
|
chr3_+_52570620
|
0.645
|
NM_001124767
|
C3orf78
|
chromosome 3 open reading frame 78
|
|
chr1_+_151763174
|
0.637
|
|
|
|
|
chr4_+_166248816
|
0.633
|
NM_001017369
NM_006745
|
MSMO1
|
methylsterol monooxygenase 1
|
|
chr17_+_4981721
|
0.629
|
NM_153018
|
ZFP3
|
zinc finger protein 3 homolog (mouse)
|
|
chr7_+_92158086
|
0.627
|
NM_032120
|
RBM48
|
RNA binding motif protein 48
|
|
chr11_+_17298278
|
0.624
|
NM_005013
|
NUCB2
|
nucleobindin 2
|
|
chr17_+_63133501
|
0.620
|
|
RGS9
|
regulator of G-protein signaling 9
|
|
chrX_-_62975005
|
0.616
|
NM_001173480
NM_015185
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chr3_+_8543539
|
0.615
|
|
LMCD1
|
LIM and cysteine-rich domains 1
|
|
chr2_+_191745546
|
0.593
|
NM_014905
|
GLS
|
glutaminase
|
|
chr17_-_7531107
|
0.590
|
NM_133491
|
SAT2
|
spermidine/spermine N1-acetyltransferase family member 2
|
|
chr7_-_148581403
|
0.590
|
NM_001203247
NM_001203248
NM_004456
NM_152998
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr1_+_212475147
|
0.589
|
NM_001199756
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha
|
|
chr6_-_41040160
|
0.588
|
NM_145063
|
C6orf130
|
chromosome 6 open reading frame 130
|
|
chr1_+_162760491
|
0.587
|
NM_016371
|
HSD17B7
|
hydroxysteroid (17-beta) dehydrogenase 7
|
|
chr1_+_82266081
|
0.584
|
NM_012302
|
LPHN2
|
latrophilin 2
|
|
chr19_+_55477710
|
0.582
|
NM_001174082
NM_001174083
NM_017852
|
NLRP2
|
NLR family, pyrin domain containing 2
|
|
chr19_+_1249882
|
0.581
|
|
MIDN
|
midnolin
|
|
chr8_+_73921061
|
0.578
|
NM_003218
NM_017489
|
TERF1
|
telomeric repeat binding factor (NIMA-interacting) 1
|
|
chr17_+_62503184
|
0.575
|
|
CEP95
|
centrosomal protein 95kDa
|
|
chr20_+_60813577
|
0.575
|
NM_014835
NM_144498
|
OSBPL2
|
oxysterol binding protein-like 2
|
|
chr20_-_45985411
|
0.575
|
NM_012408
NM_183047
NM_183048
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr1_+_1981902
|
0.571
|
NM_002744
|
PRKCZ
|
protein kinase C, zeta
|
|
chr17_+_62503079
|
0.570
|
NM_138363
|
CEP95
|
centrosomal protein 95kDa
|
|
chr4_-_89744154
|
0.569
|
NM_001015045
|
FAM13A
|
family with sequence similarity 13, member A
|
|
chr12_+_123868705
|
0.569
|
|
SETD8
|
SET domain containing (lysine methyltransferase) 8
|
|
chr10_-_77161424
|
0.566
|
NM_032772
|
ZNF503
|
zinc finger protein 503
|
|
chr1_+_26485510
|
0.564
|
NM_024869
|
FAM110D
|
family with sequence similarity 110, member D
|
|
chr19_+_21541734
|
0.563
|
|
ZNF738
|
zinc finger protein 738
|
|
chr19_+_4909447
|
0.563
|
NM_001048201
|
UHRF1
|
ubiquitin-like with PHD and ring finger domains 1
|
|
chr19_+_17581276
|
0.563
|
NM_198580
|
SLC27A1
|
solute carrier family 27 (fatty acid transporter), member 1
|
|
chr2_+_113033146
|
0.561
|
NM_198581
|
ZC3H6
|
zinc finger CCCH-type containing 6
|
|
chr9_+_131843378
|
0.556
|
NM_001135917
NM_020438
|
DOLPP1
|
dolichyl pyrophosphate phosphatase 1
|
|
chr4_+_88529680
|
0.556
|
NM_014208
|
DSPP
|
dentin sialophosphoprotein
|
|
chr19_-_14629235
|
0.552
|
|
|
|
|
chr12_+_124155649
|
0.552
|
NM_001143850
NM_024809
|
TCTN2
|
tectonic family member 2
|
|
chr18_+_60190657
|
0.546
|
NM_017742
|
ZCCHC2
|
zinc finger, CCHC domain containing 2
|
|
chr7_+_130131172
|
0.543
|
NM_177525
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr2_+_12856813
|
0.541
|
NM_021643
|
TRIB2
|
tribbles homolog 2 (Drosophila)
|
|
chr19_+_41284161
|
0.540
|
|
RAB4B-EGLN2
RAB4B
|
RAB4B-EGLN2 readthrough
RAB4B, member RAS oncogene family
|
|
chr1_+_145438437
|
0.539
|
NM_006472
|
TXNIP
|
thioredoxin interacting protein
|
|
chr8_+_68864341
|
0.539
|
|
PREX2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2
|
|
chr17_-_5095131
|
0.534
|
NM_032530
|
ZNF594
|
zinc finger protein 594
|
|
chr2_+_61293038
|
0.531
|
|
KIAA1841
|
KIAA1841
|
|
chr7_+_77325729
|
0.530
|
NM_198467
|
RSBN1L
|
round spermatid basic protein 1-like
|