|
chr8_-_22549901
|
3.002
|
NM_001199880
|
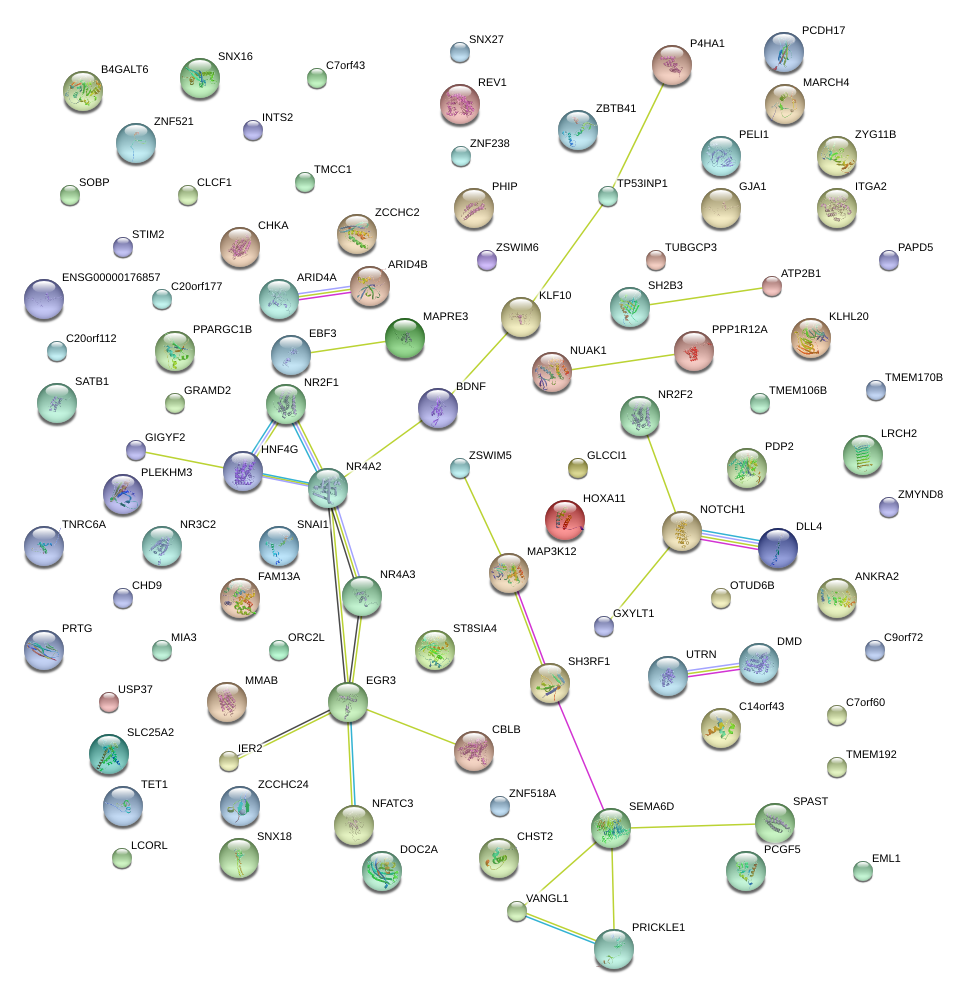
EGR3
|
early growth response 3
|
|
chr8_-_22550057
|
2.967
|
NM_001199881
|
EGR3
|
early growth response 3
|
|
chr8_-_22550708
|
2.950
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr20_+_58515408
|
2.727
|
NM_001190827
NM_022106
|
C20orf177
|
chromosome 20 open reading frame 177
|
|
chr20_+_58508818
|
2.713
|
NM_001190826
|
C20orf177
|
chromosome 20 open reading frame 177
|
|
chr12_-_42877785
|
2.705
|
NM_001144882
NM_001144883
|
PRICKLE1
|
prickle homolog 1 (Drosophila)
|
|
chr12_-_42983412
|
2.699
|
NM_153026
|
PRICKLE1
|
prickle homolog 1 (Drosophila)
|
|
chr13_+_58205788
|
2.630
|
NM_001040429
|
PCDH17
|
protocadherin 17
|
|
chr12_-_42877413
|
2.629
|
NM_001144881
|
PRICKLE1
|
prickle homolog 1 (Drosophila)
|
|
chr10_-_131762017
|
2.534
|
NM_001005463
|
EBF3
|
early B-cell factor 3
|
|
chr20_-_45985411
|
2.288
|
NM_012408
NM_183047
NM_183048
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr7_-_27224762
|
2.258
|
NM_005523
|
HOXA11
|
homeobox A11
|
|
chr4_-_149363498
|
2.079
|
NM_000901
NM_001166104
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2
|
|
chr12_-_106533810
|
2.078
|
NM_014840
|
NUAK1
|
NUAK family, SNF1-like kinase, 1
|
|
chr11_-_27681195
|
2.042
|
NM_001143816
NM_170735
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_-_27721975
|
1.983
|
NM_001143813
NM_001143814
NM_001709
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr5_-_72861376
|
1.932
|
NM_023039
|
ANKRA2
|
ankyrin repeat, family A (RFXANK-like), 2
|
|
chr11_-_27722599
|
1.838
|
NM_001143808
NM_001143809
NM_001143810
NM_001143811
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_-_27722446
|
1.837
|
NM_001143812
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_-_27743604
|
1.747
|
NM_170731
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_-_27723061
|
1.732
|
NM_170733
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_-_27721179
|
1.729
|
NM_170734
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr10_+_97889467
|
1.703
|
NM_014803
|
ZNF518A
|
zinc finger protein 518A
|
|
chr7_-_112579726
|
1.690
|
NM_152556
|
C7orf60
|
chromosome 7 open reading frame 60
|
|
chr11_-_27742224
|
1.663
|
NM_001143805
NM_001143806
NM_170732
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_-_27741192
|
1.657
|
NM_001143807
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr6_+_11538486
|
1.490
|
NM_001100829
|
TMEM170B
|
transmembrane protein 170B
|
|
chr15_-_52970808
|
1.477
|
NM_019600
|
FAM214A
|
family with sequence similarity 214, member A
|
|
chr4_+_175204827
|
1.456
|
NM_001040157
|
CEP44
|
centrosomal protein 44kDa
|
|
chr1_-_197169635
|
1.388
|
NM_194314
|
ZBTB41
|
zinc finger and BTB domain containing 41
|
|
chr1_+_116184573
|
1.353
|
NM_001172411
NM_138959
|
VANGL1
|
vang-like 1 (van gogh, Drosophila)
|
|
chr8_-_82754427
|
1.335
|
NM_022133
NM_152836
NM_152837
|
SNX16
|
sorting nexin 16
|
|
chr15_-_56035176
|
1.331
|
NM_173814
|
PRTG
|
protogenin
|
|
chr7_+_8008416
|
1.320
|
NM_138426
|
GLCCI1
|
glucocorticoid induced transcript 1
|
|
chr1_+_116185249
|
1.319
|
NM_001172412
|
VANGL1
|
vang-like 1 (van gogh, Drosophila)
|
|
chr3_-_129612371
|
1.313
|
NM_001128224
|
TMCC1
|
transmembrane and coiled-coil domain family 1
|
|
chr14_-_74253876
|
1.311
|
NM_001043318
|
C14orf43
|
chromosome 14 open reading frame 43
|
|
chr7_-_28998028
|
1.304
|
NM_014817
|
TRIL
|
TLR4 interactor with leucine-rich repeats
|
|
chr14_-_74226890
|
1.296
|
NM_194278
|
C14orf43
|
chromosome 14 open reading frame 43
|
|
chr2_-_217236666
|
1.275
|
NM_020814
|
MARCH4
|
membrane-associated ring finger (C3HC4) 4
|
|
chr8_-_103667882
|
1.247
|
NM_005655
|
KLF10
|
Kruppel-like factor 10
|
|
chr16_+_68119211
|
1.237
|
NM_004555
NM_173163
NM_173165
|
NFATC3
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3
|
|
chr6_-_119031230
|
1.218
|
NM_001178035
|
CEP85L
|
centrosomal protein 85kDa-like
|
|
chr2_-_208890207
|
1.214
|
NM_001080475
|
PLEKHM3
|
pleckstrin homology domain containing, family M, member 3
|
|
chr2_-_157189040
|
1.196
|
NM_006186
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chr2_-_219432955
|
1.193
|
NM_020935
|
USP37
|
ubiquitin specific peptidase 37
|
|
chr3_-_129599220
|
1.179
|
NM_001017395
|
TMCC1
|
transmembrane and coiled-coil domain family 1
|
|
chr14_+_58765209
|
1.172
|
NM_002892
NM_023000
NM_023001
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like)
|
|
chr12_-_42538432
|
1.152
|
NM_001099650
NM_173601
|
GXYLT1
|
glucoside xylosyltransferase 1
|
|
chr4_-_18023358
|
1.150
|
NM_001166139
NM_153686
|
LCORL
|
ligand dependent nuclear receptor corepressor-like
|
|
chr4_-_89744154
|
1.136
|
NM_001015045
|
FAM13A
|
family with sequence similarity 13, member A
|
|
chr18_+_60190657
|
1.133
|
NM_017742
|
ZCCHC2
|
zinc finger, CCHC domain containing 2
|
|
chr16_+_50186809
|
1.132
|
NM_001040284
NM_001040285
|
PAPD5
|
PAP associated domain containing 5
|
|
chr17_-_60005309
|
1.110
|
NM_020748
|
INTS2
|
integrator complex subunit 2
|
|
chr5_+_53813494
|
1.097
|
NM_001145427
NM_001102575
NM_052870
|
SNX18
|
sorting nexin 18
|
|
chr2_+_32288579
|
1.093
|
NM_014946
NM_199436
|
SPAST
|
spastin
|
|
chr8_-_103666018
|
1.092
|
NM_001032282
|
KLF10
|
Kruppel-like factor 10
|
|
chr14_+_100259445
|
1.079
|
NM_001008707
NM_004434
|
EML1
|
echinoderm microtubule associated protein like 1
|
|
chr13_-_113242467
|
1.045
|
NM_006322
|
TUBGCP3
|
tubulin, gamma complex associated protein 3
|
|
chr16_+_53088791
|
1.009
|
NM_025134
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr18_-_29264394
|
0.999
|
NM_004775
|
B4GALT6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6
|
|
chr8_-_95961543
|
0.993
|
NM_001135733
NM_033285
|
TP53INP1
|
tumor protein p53 inducible nuclear protein 1
|
|
chr2_-_64371444
|
0.983
|
NM_020651
|
PELI1
|
pellino homolog 1 (Drosophila)
|
|
chr15_+_41221530
|
0.939
|
NM_019074
|
DLL4
|
delta-like 4 (Drosophila)
|
|
chrX_-_33229428
|
0.937
|
NM_004006
|
DMD
|
dystrophin
|
|
chr16_+_66914380
|
0.927
|
NM_020786
|
PDP2
|
pyruvate dehyrogenase phosphatase catalytic subunit 2
|
|
chr10_+_70320044
|
0.918
|
NM_030625
|
TET1
|
tet methylcytosine dioxygenase 1
|
|
chr7_-_99756275
|
0.898
|
NM_018275
|
C7orf43
|
chromosome 7 open reading frame 43
|
|
chr20_-_31071187
|
0.897
|
NM_080616
|
C20orf112
|
chromosome 20 open reading frame 112
|
|
chr2_+_233561997
|
0.887
|
NM_001103146
NM_001103147
NM_001103148
NM_015575
|
GIGYF2
|
GRB10 interacting GYF protein 2
|
|
chr5_+_52284903
|
0.879
|
NM_002203
|
ITGA2
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor)
|
|
chr3_-_18466759
|
0.873
|
NM_001195470
NM_002971
|
SATB1
|
SATB homeobox 1
|
|
chr10_-_81205091
|
0.869
|
NM_153367
|
ZCCHC24
|
zinc finger, CCHC domain containing 24
|
|
chr7_+_12250818
|
0.864
|
NM_001134232
NM_018374
|
TMEM106B
|
transmembrane protein 106B
|
|
chr2_-_201828180
|
0.860
|
NM_006190
|
ORC2
|
origin recognition complex, subunit 2
|
|
chr11_-_67888857
|
0.854
|
NM_001277
NM_212469
|
CHKA
|
choline kinase alpha
|
|
chr8_+_92082423
|
0.847
|
NM_016023
|
OTUD6B
|
OTU domain containing 6B
|
|
chr20_+_48599511
|
0.845
|
NM_005985
|
SNAI1
|
snail homolog 1 (Drosophila)
|
|
chr16_+_24740945
|
0.841
|
NM_014494
|
TNRC6A
|
trinucleotide repeat containing 6A
|
|
chrX_-_114468604
|
0.839
|
NM_001243963
NM_020871
|
LRCH2
|
leucine-rich repeats and calponin homology (CH) domain containing 2
|
|
chrX_-_32173585
|
0.814
|
NM_004013
NM_004020
NM_004021
NM_004022
NM_004023
|
DMD
|
dystrophin
|
|
chr1_+_151584514
|
0.809
|
NM_030918
|
SNX27
|
sorting nexin family member 27
|
|
chr5_+_60628085
|
0.806
|
NM_020928
|
ZSWIM6
|
zinc finger, SWIM-type containing 6
|
|
chr6_-_79787939
|
0.796
|
NM_017934
|
PHIP
|
pleckstrin homology domain interacting protein
|
|
chrX_-_31526414
|
0.795
|
NM_004014
|
DMD
|
dystrophin
|
|
chr3_-_18480203
|
0.793
|
NM_001131010
|
SATB1
|
SATB homeobox 1
|
|
chr10_-_74856656
|
0.789
|
NM_000917
NM_001017962
NM_001142595
NM_001142596
|
P4HA1
|
prolyl 4-hydroxylase, alpha polypeptide I
|
|
chr12_-_80329234
|
0.786
|
NM_001143885
NM_001244990
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|
|
chr1_+_244216506
|
0.781
|
NM_006352
|
ZNF238
|
zinc finger protein 238
|
|
chr18_-_22932109
|
0.769
|
NM_015461
|
ZNF521
|
zinc finger protein 521
|
|
chr6_+_121756744
|
0.769
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr9_-_27573444
|
0.769
|
NM_018325
|
C9orf72
|
chromosome 9 open reading frame 72
|
|
chr5_+_149151502
|
0.761
|
NM_001172699
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr4_-_170192193
|
0.759
|
NM_020870
|
SH3RF1
|
SH3 domain containing ring finger 1
|
|
chr1_+_244214552
|
0.757
|
NM_205768
|
ZNF238
|
zinc finger protein 238
|
|
chrX_-_33146543
|
0.747
|
NM_004010
NM_004009
|
DMD
|
dystrophin
|
|
chr10_+_92980322
|
0.735
|
NM_032373
|
PCGF5
|
polycomb group ring finger 5
|
|
chr6_+_107811272
|
0.734
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr15_+_47476225
|
0.732
|
NM_001198999
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chrX_-_32430370
|
0.731
|
NM_004011
NM_004012
|
DMD
|
dystrophin
|
|
chr11_-_67141008
|
0.726
|
NM_013246
|
CLCF1
|
cardiotrophin-like cytokine factor 1
|
|
chr1_+_53192021
|
0.725
|
NM_024646
|
ZYG11B
|
zyg-11 homolog B (C. elegans)
|
|
chr15_+_96876568
|
0.724
|
NM_001145157
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr11_-_67141647
|
0.724
|
NM_001166212
|
CLCF1
|
cardiotrophin-like cytokine factor 1
|
|
chr12_-_80307690
|
0.720
|
NM_001143886
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|
|
chr1_-_45672225
|
0.719
|
NM_020883
|
ZSWIM5
|
zinc finger, SWIM-type containing 5
|
|
chr3_+_142838596
|
0.717
|
NM_004267
|
CHST2
|
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2
|
|
chrX_-_33357725
|
0.714
|
NM_000109
|
DMD
|
dystrophin
|
|
chr3_-_105587710
|
0.710
|
NM_170662
|
CBLB
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence b
|
|
chr1_+_173684061
|
0.706
|
NM_014458
|
KLHL20
|
kelch-like 20 (Drosophila)
|
|
chr12_-_53893195
|
0.703
|
NM_001193511
NM_006301
|
MAP3K12
|
mitogen-activated protein kinase kinase kinase 12
|
|
chr12_+_111843743
|
0.700
|
NM_005475
|
SH2B3
|
SH2B adaptor protein 3
|
|
chr9_-_139440237
|
0.693
|
NM_017617
|
NOTCH1
|
notch 1
|
|
chr1_+_222791151
|
0.693
|
NM_198551
|
MIA3
|
melanoma inhibitory activity family, member 3
|
|
chr4_+_26862312
|
0.690
|
NM_001169117
NM_001169118
NM_020860
|
STIM2
|
stromal interaction molecule 2
|
|
chr4_-_166034003
|
0.688
|
NM_001100389
|
TMEM192
|
transmembrane protein 192
|
|
chr5_-_100238870
|
0.687
|
NM_175052
NM_005668
|
ST8SIA4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4
|
|
chrX_-_31284946
|
0.682
|
NM_004015
NM_004016
NM_004017
NM_004018
NM_004019
|
DMD
|
dystrophin
|
|
chr15_-_72490109
|
0.682
|
NM_001012642
|
GRAMD2
|
GRAM domain containing 2
|
|
chr8_+_76452202
|
0.682
|
NM_004133
|
HNF4G
|
hepatocyte nuclear factor 4, gamma
|
|
chr2_-_100106478
|
0.675
|
NM_001037872
NM_016316
|
REV1
|
REV1 homolog (S. cerevisiae)
|
|
chr16_-_30022300
|
0.674
|
NM_003586
|
DOC2A
|
double C2-like domains, alpha
|
|
chr12_-_80328926
|
0.665
|
NM_002480
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|
|
chr12_-_90049818
|
0.663
|
NM_001001323
NM_001682
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1
|
|
chr19_+_13261272
|
0.663
|
NM_004907
|
IER2
|
immediate early response 2
|
|
chr2_-_99552677
|
0.659
|
NM_207362
|
C2orf55
|
chromosome 2 open reading frame 55
|
|
chr9_-_125590809
|
0.656
|
NM_005388
|
PDCL
|
phosducin-like
|
|
chr6_+_158957467
|
0.654
|
NM_020823
|
TMEM181
|
transmembrane protein 181
|
|
chr22_+_40440664
|
0.654
|
NM_001024843
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr17_-_71228482
|
0.653
|
NM_001098832
NM_032837
|
FAM104A
|
family with sequence similarity 104, member A
|
|
chr10_-_23003443
|
0.649
|
NM_005028
|
PIP4K2A
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha
|
|
chr14_+_96846021
|
0.648
|
NM_016472
|
C14orf129
|
chromosome 14 open reading frame 129
|
|
chr3_-_195163575
|
0.643
|
NM_012287
|
ACAP2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2
|
|
chr2_+_86668270
|
0.639
|
NM_018433
|
KDM3A
|
lysine (K)-specific demethylase 3A
|
|
chr22_-_22863504
|
0.637
|
NM_080764
|
ZNF280B
|
zinc finger protein 280B
|
|
chr9_-_14693459
|
0.635
|
NM_178566
|
ZDHHC21
|
zinc finger, DHHC-type containing 21
|
|
chr17_-_65241305
|
0.632
|
NM_014877
|
HELZ
|
helicase with zinc finger
|
|
chr2_-_61245363
|
0.632
|
NM_144709
|
PUS10
|
pseudouridylate synthase 10
|
|
chr5_+_149109814
|
0.630
|
NM_001172698
NM_133263
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chrX_-_72299224
|
0.627
|
NM_001012977
|
PABPC1L2A
PABPC1L2B
|
poly(A) binding protein, cytoplasmic 1-like 2A
poly(A) binding protein, cytoplasmic 1-like 2B
|
|
chr17_-_76356153
|
0.627
|
NM_003955
|
SOCS3
|
suppressor of cytokine signaling 3
|
|
chr1_+_204042191
|
0.626
|
NM_005686
|
SOX13
|
SRY (sex determining region Y)-box 13
|
|
chr17_+_76000248
|
0.624
|
NM_001142640
NM_018996
|
TNRC6C
|
trinucleotide repeat containing 6C
|
|
chr15_+_96875793
|
0.622
|
NM_001145156
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr1_+_64936475
|
0.617
|
NM_020925
|
CACHD1
|
cache domain containing 1
|
|
chr5_-_141392537
|
0.617
|
NM_005471
|
GNPDA1
|
glucosamine-6-phosphate deaminase 1
|
|
chr4_+_174089889
|
0.611
|
NM_017423
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7)
|
|
chr15_+_51200864
|
0.610
|
NM_001252127
NM_007347
|
AP4E1
|
adaptor-related protein complex 4, epsilon 1 subunit
|
|
chr1_-_236030219
|
0.608
|
NM_000081
|
LYST
|
lysosomal trafficking regulator
|
|
chr14_+_53196852
|
0.607
|
NM_001130701
NM_145251
|
STYX
|
serine/threonine/tyrosine interacting protein
|
|
chr15_-_43662107
|
0.596
|
NM_152455
|
ZSCAN29
|
zinc finger and SCAN domain containing 29
|
|
chr7_+_21467572
|
0.596
|
NM_003112
|
SP4
|
Sp4 transcription factor
|
|
chr6_-_53409897
|
0.595
|
NM_001197115
NM_001498
|
GCLC
|
glutamate-cysteine ligase, catalytic subunit
|
|
chr5_+_89770680
|
0.594
|
NM_006467
|
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD)
|
|
chr3_+_107241782
|
0.592
|
NM_001142568
NM_020235
|
BBX
|
bobby sox homolog (Drosophila)
|
|
chr17_-_61920328
|
0.584
|
NM_001098426
|
SMARCD2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2
|
|
chr8_+_124780742
|
0.584
|
NM_144963
|
FAM91A1
|
family with sequence similarity 91, member A1
|
|
chr17_-_2304217
|
0.583
|
NM_020310
|
MNT
|
MAX binding protein
|
|
chr6_-_118973019
|
0.582
|
NM_001042475
NM_206921
|
CEP85L
|
centrosomal protein 85kDa-like
|
|
chr2_-_86564632
|
0.577
|
NM_001164731
NM_001164732
NM_022912
|
REEP1
|
receptor accessory protein 1
|
|
chr6_+_15246299
|
0.575
|
NM_004973
|
JARID2
|
jumonji, AT rich interactive domain 2
|
|
chr17_-_37557851
|
0.573
|
NM_001184906
NM_032875
|
FBXL20
|
F-box and leucine-rich repeat protein 20
|
|
chr3_-_50605062
|
0.571
|
NM_001171740
NM_001171743
NM_016210
|
C3orf18
|
chromosome 3 open reading frame 18
|
|
chr8_+_26240522
|
0.569
|
NM_004331
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like
|
|
chr13_-_73301726
|
0.568
|
NM_001071775
|
MZT1
|
mitotic spindle organizing protein 1
|
|
chr15_+_96869156
|
0.567
|
NM_001145155
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr9_+_131799212
|
0.567
|
NM_032809
|
FAM73B
|
family with sequence similarity 73, member B
|
|
chr19_+_925984
|
0.566
|
NM_005224
|
ARID3A
|
AT rich interactive domain 3A (BRIGHT-like)
|
|
chr17_+_4981721
|
0.564
|
NM_153018
|
ZFP3
|
zinc finger protein 3 homolog (mouse)
|
|
chr1_-_44497132
|
0.564
|
NM_001024845
|
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9
|
|
chr1_+_65210724
|
0.560
|
NM_018211
|
RAVER2
|
ribonucleoprotein, PTB-binding 2
|
|
chr5_+_176560056
|
0.556
|
NM_172349
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr3_-_125094003
|
0.554
|
NM_021964
|
ZNF148
|
zinc finger protein 148
|
|
chr6_-_99963245
|
0.554
|
NM_001080481
|
USP45
|
ubiquitin specific peptidase 45
|
|
chr9_+_4679557
|
0.552
|
NM_017913
|
CDC37L1
|
cell division cycle 37 homolog (S. cerevisiae)-like 1
|
|
chrX_+_107069081
|
0.551
|
NM_012216
NM_052817
|
MID2
|
midline 2
|
|
chr12_-_31743822
|
0.551
|
NM_144973
|
DENND5B
|
DENN/MADD domain containing 5B
|
|
chr7_+_26331493
|
0.550
|
NM_001199835
NM_013322
|
SNX10
|
sorting nexin 10
|
|
chr1_-_179112178
|
0.550
|
NM_001136000
NM_001168239
NM_005158
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr12_+_69633316
|
0.546
|
NM_007007
|
CPSF6
|
cleavage and polyadenylation specific factor 6, 68kDa
|
|
chr12_-_12419702
|
0.545
|
NM_002336
|
LRP6
|
low density lipoprotein receptor-related protein 6
|
|
chr15_-_56535475
|
0.542
|
NM_022841
|
RFX7
|
regulatory factor X, 7
|
|
chr12_-_39299335
|
0.541
|
NM_153634
|
CPNE8
|
copine VIII
|
|
chr2_-_70475563
|
0.540
|
NM_022037
NM_022173
|
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein
|
|
chrX_+_72782961
|
0.539
|
NM_001039840
|
CHIC1
|
cysteine-rich hydrophobic domain 1
|
|
chr5_+_174905513
|
0.538
|
NM_022754
|
SFXN1
|
sideroflexin 1
|
|
chr2_-_169103886
|
0.538
|
NM_013233
|
STK39
|
serine threonine kinase 39
|
|
chr9_-_111882209
|
0.537
|
NM_032012
|
C9orf5
|
chromosome 9 open reading frame 5
|
|
chr15_-_65579017
|
0.536
|
NM_017851
|
PARP16
|
poly (ADP-ribose) polymerase family, member 16
|
|
chr19_-_6110561
|
0.533
|
NM_000635
NM_134433
|
RFX2
|
regulatory factor X, 2 (influences HLA class II expression)
|
|
chr5_+_86564044
|
0.530
|
NM_002890
|
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1
|
|
chr2_-_38604430
|
0.530
|
NM_001135673
NM_022374
|
ATL2
|
atlastin GTPase 2
|
|
chr9_-_138987096
|
0.526
|
NM_144653
|
NACC2
|
NACC family member 2, BEN and BTB (POZ) domain containing
|
|
chr20_+_306205
|
0.525
|
NM_006943
|
SOX12
|
SRY (sex determining region Y)-box 12
|
|
chr12_+_122516555
|
0.525
|
NM_014938
|
MLXIP
|
MLX interacting protein
|
|
chr10_-_94333847
|
0.517
|
NM_004969
|
IDE
|
insulin-degrading enzyme
|
|
chr4_-_85887543
|
0.517
|
NM_014991
|
WDFY3
|
WD repeat and FYVE domain containing 3
|
|
chrX_+_48916515
|
0.515
|
NM_001163322
NM_033626
NM_001163321
NM_001163323
|
CCDC120
|
coiled-coil domain containing 120
|
|
chr9_+_131843378
|
0.514
|
NM_001135917
NM_020438
|
DOLPP1
|
dolichyl pyrophosphate phosphatase 1
|
|
chr10_+_74033676
|
0.513
|
NM_019058
|
DDIT4
|
DNA-damage-inducible transcript 4
|