|
chr2_+_228678556
|
3.203
|
NM_001130046
NM_004591
|
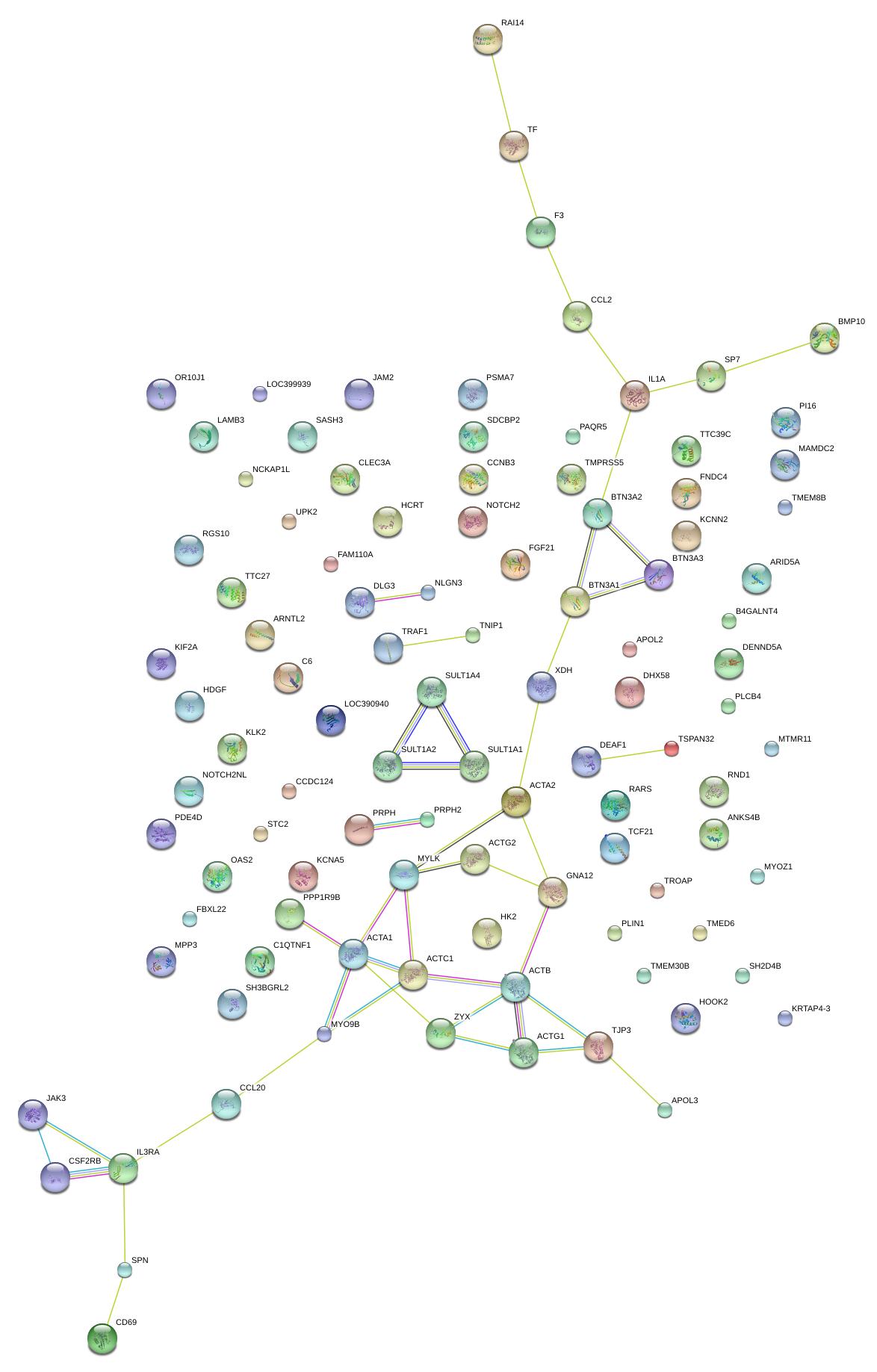
CCL20
|
chemokine (C-C motif) ligand 20
|
|
chr19_-_17958773
|
3.106
|
|
JAK3
|
Janus kinase 3
|
|
chr1_-_120612019
|
3.047
|
|
NOTCH2
|
notch 2
|
|
chr5_-_150467220
|
3.028
|
NM_001252390
|
TNIP1
|
TNFAIP3 interacting protein 1
|
|
chr22_-_36556781
|
2.958
|
NM_145640
|
APOL3
|
apolipoprotein L, 3
|
|
chr5_-_150466696
|
2.710
|
NM_001252391
NM_001252392
|
TNIP1
|
TNFAIP3 interacting protein 1
|
|
chr9_+_72658496
|
2.365
|
NM_153267
|
MAMDC2
|
MAM domain containing 2
|
|
chr1_-_149908759
|
2.326
|
NM_001145862
|
MTMR11
|
myotubularin related protein 11
|
|
chr1_-_149908721
|
1.964
|
|
MTMR11
|
myotubularin related protein 11
|
|
chr6_-_30652434
|
1.789
|
|
PPP1R18
|
protein phosphatase 1, regulatory subunit 18
|
|
chr9_+_72658735
|
1.715
|
|
MAMDC2
|
MAM domain containing 2
|
|
chr21_+_27012045
|
1.581
|
|
JAM2
|
junctional adhesion molecule 2
|
|
chr1_-_156722023
|
1.500
|
NM_001126051
NM_001126050
|
HDGF
|
hepatoma-derived growth factor
|
|
chr2_-_113542970
|
1.483
|
NM_000575
|
IL1A
|
interleukin 1, alpha
|
|
chr17_+_77020278
|
1.360
|
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1
|
|
chr3_-_123411185
|
1.301
|
|
MYLK
|
myosin light chain kinase
|
|
chrX_+_1455508
|
1.290
|
NM_002183
|
IL3RA
|
interleukin 3 receptor, alpha (low affinity)
|
|
chrY_+_1405508
|
1.245
|
NM_002183
|
IL3RA
|
interleukin 3 receptor, alpha (low affinity)
|
|
chr17_+_32582295
|
1.145
|
NM_002982
|
CCL2
|
chemokine (C-C motif) ligand 2
|
|
chr12_+_27485767
|
1.101
|
NM_001248002
NM_001248003
NM_001248004
NM_001248005
NM_020183
|
ARNTL2
|
aryl hydrocarbon receptor nuclear translocator-like 2
|
|
chr11_+_118827007
|
1.028
|
NM_006760
|
UPK2
|
uroplakin 2
|
|
chr6_+_134210247
|
1.022
|
NM_198392
NM_003206
|
TCF21
|
transcription factor 21
|
|
chr6_+_26440699
|
1.016
|
NM_001242803
NM_006994
NM_197974
|
BTN3A3
|
butyrophilin, subfamily 3, member A3
|
|
chr19_+_17186569
|
1.014
|
NM_001130065
NM_004145
|
MYO9B
|
myosin IXB
|
|
chr2_-_27718098
|
0.996
|
NM_022823
|
FNDC4
|
fibronectin type III domain containing 4
|
|
chr12_+_54891539
|
0.969
|
|
NCKAP1L
|
NCK-associated protein 1-like
|
|
chr18_+_21572736
|
0.963
|
NM_153211
|
TTC39C
|
tetratricopeptide repeat domain 39C
|
|
chr2_-_27718059
|
0.916
|
|
FNDC4
|
fibronectin type III domain containing 4
|
|
chr11_-_9225780
|
0.912
|
|
DENND5A
|
DENN/MADD domain containing 5A
|
|
chr9_-_123688352
|
0.906
|
|
TRAF1
|
TNF receptor-associated factor 1
|
|
chrX_+_128913881
|
0.896
|
NM_018990
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chrX_+_128913897
|
0.894
|
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chr14_-_61748510
|
0.885
|
NM_001017970
|
TMEM30B
|
transmembrane protein 30B
|
|
chr20_+_816710
|
0.874
|
NM_207121
|
FAM110A
|
family with sequence similarity 110, member A
|
|
chr11_+_369723
|
0.868
|
NM_178537
|
B4GALNT4
|
beta-1,4-N-acetyl-galactosaminyl transferase 4
|
|
chr12_+_113416267
|
0.862
|
NM_001032731
NM_002535
NM_016817
|
OAS2
|
2'-5'-oligoadenylate synthetase 2, 69/71kDa
|
|
chr2_+_97202463
|
0.856
|
NM_212481
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like)
|
|
chr5_+_113769226
|
0.849
|
NM_170775
|
KCNN2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2
|
|
chr6_+_26402464
|
0.845
|
NM_001145008
NM_001145009
NM_007048
NM_194441
|
BTN3A1
|
butyrophilin, subfamily 3, member A1
|
|
chr22_+_37309618
|
0.838
|
NM_000395
|
CSF2RB
|
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage)
|
|
chr17_-_40337384
|
0.836
|
NM_001524
|
HCRT
|
hypocretin (orexin) neuropeptide precursor
|
|
chr10_-_90712510
|
0.831
|
NM_001613
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr16_-_28608367
|
0.817
|
NM_001054
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2
|
|
chrX_+_70364680
|
0.809
|
NM_001166660
NM_018977
NM_181303
|
NLGN3
|
neuroligin 3
|
|
chrX_+_70364707
|
0.793
|
|
NLGN3
|
neuroligin 3
|
|
chr17_-_40264687
|
0.784
|
NM_024119
|
DHX58
|
DEXH (Asp-Glu-X-His) box polypeptide 58
|
|
chr10_-_121296044
|
0.774
|
NM_002925
|
RGS10
|
regulator of G-protein signaling 10
|
|
chr2_-_69098502
|
0.762
|
NM_014482
|
BMP10
|
bone morphogenetic protein 10
|
|
chr7_+_143079925
|
0.752
|
|
ZYX
|
zyxin
|
|
chr19_-_51893791
|
0.751
|
NM_001193623
|
LOC147646
|
uncharacterized LOC147646
|
|
chr1_-_209825797
|
0.742
|
NM_000228
|
LAMB3
|
laminin, beta 3
|
|
chrX_+_70364728
|
0.739
|
|
NLGN3
|
neuroligin 3
|
|
chr2_+_75059781
|
0.733
|
NM_000189
|
HK2
|
hexokinase 2
|
|
chr1_+_159409511
|
0.730
|
NM_012351
|
OR10J1
|
olfactory receptor, family 10, subfamily J, member 1
|
|
chr16_+_78056411
|
0.715
|
NM_005752
|
CLEC3A
|
C-type lectin domain family 3, member A
|
|
chrX_+_50027539
|
0.710
|
NM_033031
NM_033670
|
CCNB3
|
cyclin B3
|
|
chr19_+_44080951
|
0.709
|
NM_001193621
|
LOC390940
|
uncharacterized protein ENSP00000244321
|
|
chr14_-_24777064
|
0.706
|
|
|
|
|
chr3_-_123339423
|
0.706
|
NM_053031
NM_053032
|
MYLK
|
myosin light chain kinase
|
|
chr4_-_38666429
|
0.682
|
|
FLJ13197
|
uncharacterized FLJ13197
|
|
chr10_+_82297657
|
0.678
|
NM_207372
|
SH2D4B
|
SH2 domain containing 4B
|
|
chr11_+_2323356
|
0.669
|
|
TSPAN32
|
tetraspanin 32
|
|
chr1_-_209825673
|
0.648
|
NM_001127641
|
LAMB3
|
laminin, beta 3
|
|
chr15_+_69606741
|
0.643
|
NM_001104554
|
PAQR5
|
progestin and adipoQ receptor family member V
|
|
chr20_+_9288446
|
0.642
|
NM_000933
|
PLCB4
|
phospholipase C, beta 4
|
|
chr16_-_69385656
|
0.640
|
NM_144676
|
TMED6
|
transmembrane emp24 protein transport domain containing 6
|
|
chr12_+_49716970
|
0.632
|
NM_001100620
NM_005480
|
TROAP
|
trophinin associated protein (tastin)
|
|
chr20_-_1306512
|
0.630
|
NM_001199784
|
SDCBP2
|
syndecan binding protein (syntenin) 2
|
|
chr15_-_90222572
|
0.626
|
NM_001145311
NM_002666
|
PLIN1
|
perilipin 1
|
|
chr5_-_172756505
|
0.623
|
NM_003714
|
STC2
|
stanniocalcin 2
|
|
chr11_-_113577023
|
0.614
|
NM_030770
|
TMPRSS5
|
transmembrane protease, serine 5
|
|
chr10_-_75401492
|
0.606
|
NM_021245
|
MYOZ1
|
myozenin 1
|
|
chr12_-_53730003
|
0.606
|
NM_001173467
|
SP7
|
Sp7 transcription factor
|
|
chr19_+_18045904
|
0.601
|
NM_138442
|
CCDC124
|
coiled-coil domain containing 124
|
|
chr12_-_49259549
|
0.600
|
NM_014470
|
RND1
|
Rho family GTPase 1
|
|
chr5_+_34656482
|
0.590
|
|
RAI14
|
retinoic acid induced 14
|
|
chr9_+_35829375
|
0.586
|
NM_001042590
|
TMEM8B
|
transmembrane protein 8B
|
|
chr7_-_2883649
|
0.585
|
NM_007353
|
GNA12
|
guanine nucleotide binding protein (G protein) alpha 12
|
|
chr11_-_89650877
|
0.585
|
NM_001206627
|
LOC399939
|
ring finger protein 18-like
|
|
chr6_+_80340971
|
0.582
|
NM_031469
|
SH3BGRL2
|
SH3 domain binding glutamic acid-rich protein like 2
|
|
chr15_+_63889551
|
0.560
|
NM_203373
|
FBXL22
|
F-box and leucine-rich repeat protein 22
|
|
chr16_+_29674564
|
0.560
|
|
SPN
|
sialophorin
|
|
chr5_-_41261539
|
0.554
|
NM_001115131
|
C6
|
complement component 6
|
|
chr5_-_59783885
|
0.552
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr17_-_39324423
|
0.543
|
NM_033187
|
KRTAP4-3
|
keratin associated protein 4-3
|
|
chr6_+_36922208
|
0.542
|
NM_153370
|
PI16
|
peptidase inhibitor 16
|
|
chr3_+_133494275
|
0.542
|
|
TF
|
transferrin
|
|
chr5_+_167913445
|
0.540
|
NM_002887
|
RARS
|
arginyl-tRNA synthetase
|
|
chr19_+_3708358
|
0.538
|
|
TJP3
|
tight junction protein 3 (zona occludens 3)
|
|
chr2_+_32853086
|
0.537
|
NM_001193509
NM_017735
|
TTC27
|
tetratricopeptide repeat domain 27
|
|
chr12_-_9913415
|
0.530
|
NM_001781
|
CD69
|
CD69 molecule
|
|
chr11_+_2323236
|
0.525
|
NM_139022
|
TSPAN32
|
tetraspanin 32
|
|
chr16_+_21245015
|
0.525
|
NM_145865
|
ANKS4B
|
ankyrin repeat and sterile alpha motif domain containing 4B
|
|
chr19_+_49259343
|
0.519
|
NM_019113
|
FGF21
|
fibroblast growth factor 21
|
|
chr2_-_31637553
|
0.515
|
|
XDH
|
xanthine dehydrogenase
|
|
chr12_+_49688908
|
0.515
|
NM_006262
|
PRPH
|
peripherin
|
|
chrX_+_69672356
|
0.514
|
|
DLG3
|
discs, large homolog 3 (Drosophila)
|
|
chr11_+_89659929
|
0.511
|
NM_001206627
|
LOC399939
|
ring finger protein 18-like
|
|
chr7_+_99816870
|
0.506
|
NM_024070
|
PVRIG
|
poliovirus receptor related immunoglobulin domain containing
|
|
chr12_+_54891522
|
0.502
|
|
NCKAP1L
|
NCK-associated protein 1-like
|
|
chrX_+_49832214
|
0.501
|
NM_000084
|
CLCN5
|
chloride channel 5
|
|
chr10_+_124145584
|
0.500
|
NM_001195608
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chr12_+_75874512
|
0.499
|
NM_006851
|
GLIPR1
|
GLI pathogenesis-related 1
|
|
chr17_-_39646115
|
0.494
|
NM_003771
|
KRT36
|
keratin 36
|
|
chr1_-_173174611
|
0.483
|
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4
|
|
chr6_-_39282095
|
0.482
|
NM_031460
NM_001135111
|
KCNK17
|
potassium channel, subfamily K, member 17
|
|
chr6_+_11094265
|
0.479
|
NM_001135575
|
C6orf228
|
chromosome 6 open reading frame 228
|
|
chr1_-_26644755
|
0.478
|
NM_145345
|
UBXN11
|
UBX domain protein 11
|
|
chr3_-_123339178
|
0.477
|
|
MYLK
|
myosin light chain kinase
|
|
chr7_-_76038894
|
0.475
|
NM_080744
|
SRCRB4D
|
scavenger receptor cysteine rich domain containing, group B (4 domains)
|
|
chr22_+_31478845
|
0.475
|
NM_001207017
|
SMTN
|
smoothelin
|
|
chr3_+_148415657
|
0.474
|
NM_000685
NM_004835
NM_009585
NM_031850
|
AGTR1
|
angiotensin II receptor, type 1
|
|
chr1_-_54199876
|
0.474
|
NM_147193
|
GLIS1
|
GLIS family zinc finger 1
|
|
chr7_-_141957846
|
0.473
|
NM_001001317
|
PRSS58
|
protease, serine, 58
|
|
chr5_-_138862342
|
0.470
|
NM_198282
|
TMEM173
|
transmembrane protein 173
|
|
chr21_+_37507209
|
0.469
|
NM_001236
|
CBR3
|
carbonyl reductase 3
|
|
chr11_-_117800067
|
0.467
|
NM_001077263
NM_001206789
NM_001206790
NM_001244995
|
TMPRSS13
|
transmembrane protease, serine 13
|
|
chr7_-_75419063
|
0.461
|
NM_006072
|
CCL26
|
chemokine (C-C motif) ligand 26
|
|
chr4_-_40631846
|
0.459
|
NM_001098634
|
RBM47
|
RNA binding motif protein 47
|
|
chr1_+_202091971
|
0.458
|
NM_004767
|
GPR37L1
|
G protein-coupled receptor 37 like 1
|
|
chr12_+_26205490
|
0.457
|
NM_001164746
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chr1_-_173446273
|
0.453
|
|
LOC100506023
|
uncharacterized LOC100506023
|
|
chr6_+_134210264
|
0.453
|
|
TCF21
|
transcription factor 21
|
|
chr19_-_42931500
|
0.452
|
NM_005357
|
LIPE
|
lipase, hormone-sensitive
|
|
chr11_-_64703334
|
0.449
|
NM_130769
|
GPHA2
|
glycoprotein hormone alpha 2
|
|
chr5_-_138862224
|
0.440
|
|
TMEM173
|
transmembrane protein 173
|
|
chr1_+_11249344
|
0.436
|
NM_021146
|
ANGPTL7
|
angiopoietin-like 7
|
|
chr3_-_195310985
|
0.433
|
NM_001647
|
APOD
|
apolipoprotein D
|
|
chr20_+_58296264
|
0.431
|
NM_183244
NM_183246
|
PHACTR3
|
phosphatase and actin regulator 3
|
|
chr4_-_40632639
|
0.430
|
|
RBM47
|
RNA binding motif protein 47
|
|
chr3_-_190167597
|
0.429
|
NM_207316
|
TMEM207
|
transmembrane protein 207
|
|
chr14_+_74083547
|
0.427
|
NM_001037162
|
ACOT6
|
acyl-CoA thioesterase 6
|
|
chr10_-_99531712
|
0.427
|
NM_003015
|
SFRP5
|
secreted frizzled-related protein 5
|
|
chr3_+_149192367
|
0.426
|
NM_004617
|
TM4SF4
|
transmembrane 4 L six family member 4
|
|
chr5_-_146460991
|
0.426
|
NM_181677
NM_181678
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta
|
|
chr10_-_104211244
|
0.422
|
NM_024886
|
C10orf95
|
chromosome 10 open reading frame 95
|
|
chr2_-_128100673
|
0.420
|
NM_006609
|
MAP3K2
|
mitogen-activated protein kinase kinase kinase 2
|
|
chr16_+_57438678
|
0.420
|
NM_002987
|
CCL17
|
chemokine (C-C motif) ligand 17
|
|
chr10_+_71812356
|
0.420
|
NM_018649
|
H2AFY2
|
H2A histone family, member Y2
|
|
chr17_+_3118911
|
0.419
|
NM_014565
|
OR1A1
|
olfactory receptor, family 1, subfamily A, member 1
|
|
chr9_+_35829208
|
0.419
|
NM_016446
NM_001042589
|
TMEM8B
|
transmembrane protein 8B
|
|
chr12_-_91505541
|
0.416
|
NM_002345
|
LUM
|
lumican
|
|
chr12_+_41086189
|
0.415
|
NM_001843
NM_175038
|
CNTN1
|
contactin 1
|
|
chr3_-_195310770
|
0.415
|
|
APOD
|
apolipoprotein D
|
|
chr1_+_36636690
|
0.415
|
|
MAP7D1
|
MAP7 domain containing 1
|
|
chr17_+_73720775
|
0.411
|
NM_001005619
|
ITGB4
|
integrin, beta 4
|
|
chr2_+_102803432
|
0.410
|
NM_003854
|
IL1RL2
|
interleukin 1 receptor-like 2
|
|
chr6_-_114384008
|
0.409
|
NM_153612
|
HS3ST5
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 5
|
|
chr11_-_57004674
|
0.409
|
NM_005161
|
APLNR
|
apelin receptor
|
|
chr6_-_161087406
|
0.407
|
NM_005577
|
LPA
|
lipoprotein, Lp(a)
|
|
chr11_-_64764388
|
0.406
|
NM_138456
|
BATF2
|
basic leucine zipper transcription factor, ATF-like 2
|
|
chr17_-_39306053
|
0.405
|
NM_033188
|
KRTAP4-5
|
keratin associated protein 4-5
|
|
chr1_+_32661018
|
0.403
|
|
TXLNA
|
taxilin alpha
|
|
chr2_-_100939194
|
0.403
|
NM_198461
|
LONRF2
|
LON peptidase N-terminal domain and ring finger 2
|
|
chrX_-_69479516
|
0.402
|
NM_002565
|
P2RY4
|
pyrimidinergic receptor P2Y, G-protein coupled, 4
|
|
chr7_-_20256942
|
0.401
|
NM_182762
|
MACC1
|
metastasis associated in colon cancer 1
|
|
chr2_-_85637486
|
0.400
|
NM_001747
|
CAPG
|
capping protein (actin filament), gelsolin-like
|
|
chr1_-_209824641
|
0.400
|
NM_001017402
|
LAMB3
|
laminin, beta 3
|
|
chr14_-_69259654
|
0.400
|
|
ZFP36L1
|
zinc finger protein 36, C3H type-like 1
|
|
chr20_+_123230
|
0.394
|
NM_030931
|
DEFB126
|
defensin, beta 126
|
|
chr5_+_34687663
|
0.393
|
NM_001145523
|
RAI14
|
retinoic acid induced 14
|
|
chr9_-_25678855
|
0.391
|
NM_001004125
|
TUSC1
|
tumor suppressor candidate 1
|
|
chr20_-_42355566
|
0.388
|
NM_001008901
NM_176791
|
GTSF1L
|
gametocyte specific factor 1-like
|
|
chr19_+_47840370
|
0.379
|
NM_018485
|
GPR77
|
G protein-coupled receptor 77
|
|
chr7_-_84751246
|
0.378
|
NM_152754
|
SEMA3D
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D
|
|
chr6_+_167525294
|
0.373
|
NM_004367
|
CCR6
|
chemokine (C-C motif) receptor 6
|
|
chr3_+_149192479
|
0.372
|
|
TM4SF4
|
transmembrane 4 L six family member 4
|
|
chr11_+_71903172
|
0.362
|
NM_016729
|
FOLR1
|
folate receptor 1 (adult)
|
|
chr4_+_103422479
|
0.362
|
NM_001165412
NM_003998
|
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1
|
|
chr21_+_34602205
|
0.362
|
NM_000874
NM_207584
NM_207585
|
IFNAR2
|
interferon (alpha, beta and omega) receptor 2
|
|
chr17_+_16319087
|
0.361
|
|
TRPV2
|
transient receptor potential cation channel, subfamily V, member 2
|
|
chr18_+_43405544
|
0.360
|
NM_213602
|
SIGLEC15
|
sialic acid binding Ig-like lectin 15
|
|
chr16_-_4854748
|
0.360
|
|
GLYR1
|
glyoxylate reductase 1 homolog (Arabidopsis)
|
|
chr11_-_119999516
|
0.358
|
|
TRIM29
|
tripartite motif containing 29
|
|
chr8_-_125580745
|
0.358
|
|
MTSS1
|
metastasis suppressor 1
|
|
chr21_-_43771069
|
0.357
|
NM_005423
|
TFF2
|
trefoil factor 2
|
|
chr1_+_44444848
|
0.356
|
NM_003780
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2
|
|
chr17_-_8770976
|
0.354
|
NM_001010855
|
PIK3R6
|
phosphoinositide-3-kinase, regulatory subunit 6
|
|
chr1_-_183274008
|
0.353
|
NM_170706
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2
|
|
chr19_+_55587220
|
0.352
|
NM_133180
|
EPS8L1
|
EPS8-like 1
|
|
chr5_-_160279045
|
0.351
|
NM_025153
|
ATP10B
|
ATPase, class V, type 10B
|
|
chr5_-_149792370
|
0.347
|
NM_001025158
NM_001025159
NM_004355
|
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain
|
|
chr22_+_41957011
|
0.347
|
NM_014460
|
CSDC2
|
cold shock domain containing C2, RNA binding
|
|
chr11_-_117698727
|
0.346
|
NM_021603
|
FXYD2
|
FXYD domain containing ion transport regulator 2
|
|
chr4_-_95263986
|
0.345
|
NM_014485
|
HPGDS
|
hematopoietic prostaglandin D synthase
|
|
chr11_-_10673832
|
0.345
|
NM_001098579
|
MRVI1
|
murine retrovirus integration site 1 homolog
|
|
chrX_+_47050208
|
0.341
|
|
UBA1
|
ubiquitin-like modifier activating enzyme 1
|
|
chr11_+_60869929
|
0.340
|
NM_014207
|
CD5
|
CD5 molecule
|
|
chr9_-_72873739
|
0.339
|
|
LOC100507299
|
uncharacterized LOC100507299
|
|
chr14_+_70078313
|
0.339
|
|
KIAA0247
|
KIAA0247
|
|
chr5_+_149887673
|
0.338
|
NM_001543
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1
|
|
chr11_-_64527369
|
0.338
|
|
PYGM
|
phosphorylase, glycogen, muscle
|
|
chr17_+_18380097
|
0.337
|
NM_001040078
|
LGALS9B
LGALS9C
|
lectin, galactoside-binding, soluble, 9B
lectin, galactoside-binding, soluble, 9C
|
|
chr2_-_228497801
|
0.337
|
NM_001162483
NM_020161
|
C2orf83
|
chromosome 2 open reading frame 83
|
|
chr16_-_20338808
|
0.336
|
NM_001007240
NM_001007241
NM_001007242
NM_001502
|
GP2
|
glycoprotein 2 (zymogen granule membrane)
|
|
chr12_-_53600999
|
0.336
|
NM_000889
|
ITGB7
|
integrin, beta 7
|
|
chr4_+_47033294
|
0.336
|
NM_000812
|
GABRB1
|
gamma-aminobutyric acid (GABA) A receptor, beta 1
|
|
chr10_-_101380160
|
0.335
|
|
SLC25A28
|
solute carrier family 25, member 28
|
|
chr11_-_65667809
|
0.335
|
|
FOSL1
|
FOS-like antigen 1
|
|
chr3_-_71542628
|
0.335
|
|
FOXP1
|
forkhead box P1
|