|
chr2_-_7005764
|
7.819
|
NM_207315
|
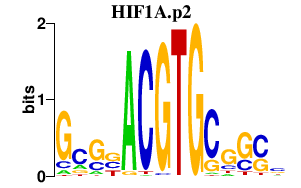
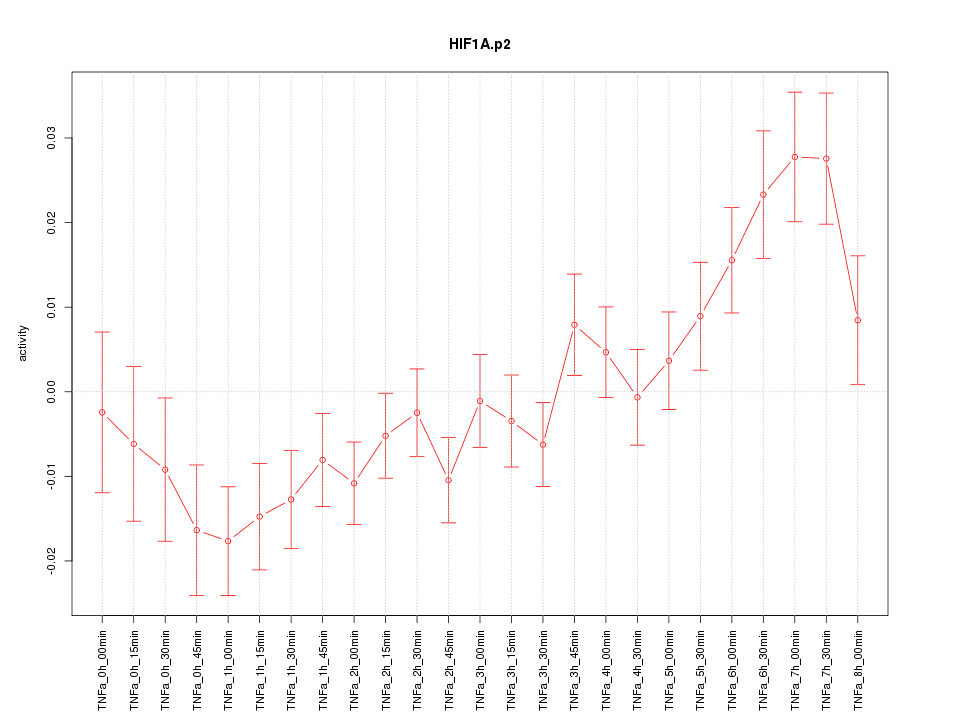
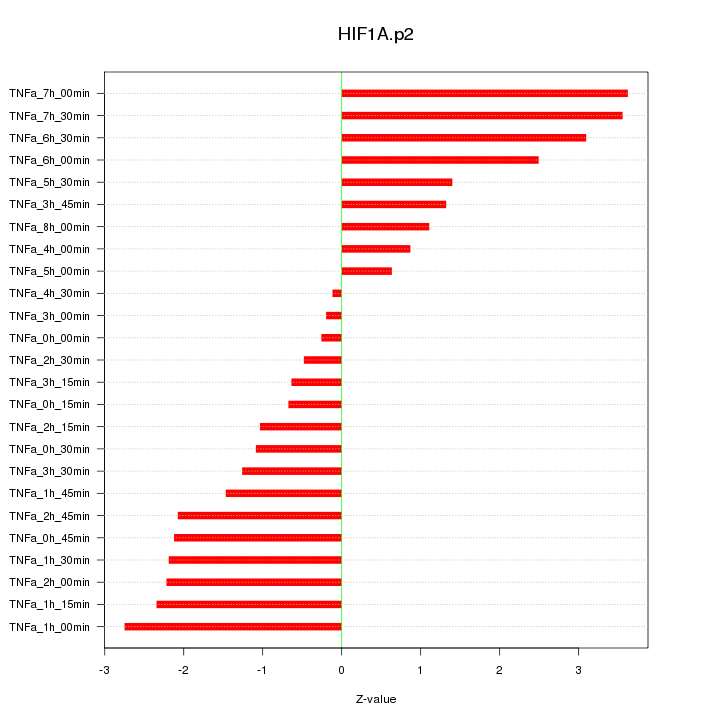
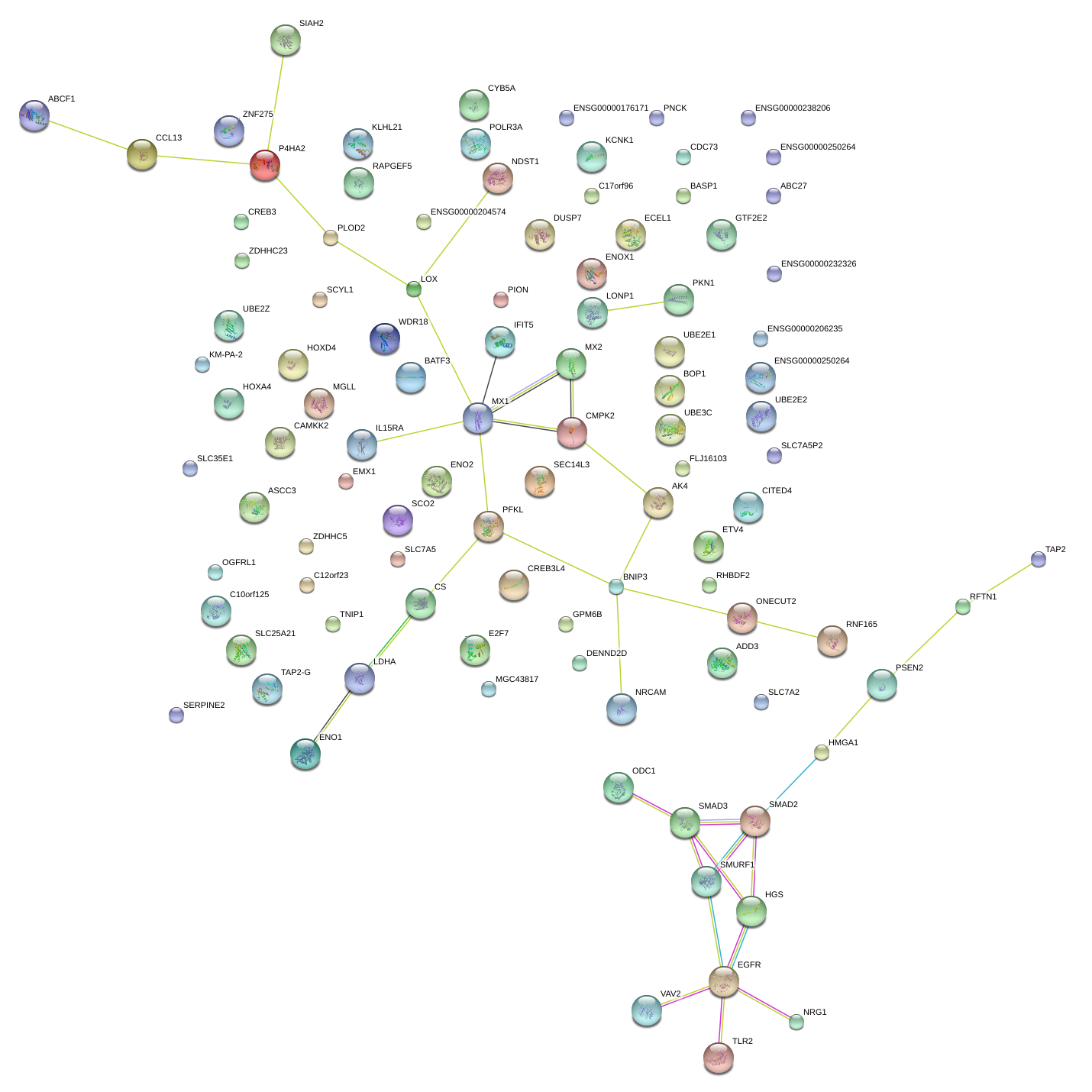
CMPK2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial
|
|
chr1_+_65613211
|
7.163
|
NM_203464
NM_001005353
|
AK4
|
adenylate kinase 4
|
|
chr1_+_65613971
|
7.094
|
|
AK4
|
adenylate kinase 4
|
|
chr3_-_127541160
|
5.334
|
NM_007283
|
MGLL
|
monoglyceride lipase
|
|
chr3_-_127541975
|
5.267
|
NM_001003794
|
MGLL
|
monoglyceride lipase
|
|
chr8_+_17354562
|
5.033
|
NM_001008539
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr17_-_74497406
|
4.963
|
NM_024599
|
RHBDF2
|
rhomboid 5 homolog 2 (Drosophila)
|
|
chr1_+_65613771
|
4.674
|
NM_013410
|
AK4
|
adenylate kinase 4
|
|
chr8_+_31497267
|
4.173
|
NM_013962
|
NRG1
|
neuregulin 1
|
|
chr3_-_145879222
|
4.145
|
NM_000935
NM_182943
|
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2
|
|
chr7_+_55086922
|
3.948
|
|
EGFR
|
epidermal growth factor receptor
|
|
chr10_-_6019454
|
3.946
|
NM_002189
NM_172200
|
IL15RA
|
interleukin 15 receptor, alpha
|
|
chr11_+_18416111
|
3.808
|
|
LDHA
|
lactate dehydrogenase A
|
|
chr11_+_18416106
|
3.747
|
|
LDHA
|
lactate dehydrogenase A
|
|
chr11_+_18415917
|
3.399
|
NM_001135239
NM_001165415
NM_001165416
NM_005566
|
LDHA
|
lactate dehydrogenase A
|
|
chr13_-_44361032
|
3.356
|
NM_001242863
NM_017993
|
ENOX1
|
ecto-NOX disulfide-thiol exchanger 1
|
|
chr3_-_145878936
|
3.250
|
|
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2
|
|
chr5_-_131563481
|
3.149
|
NM_001017974
NM_001142598
NM_001142599
|
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II
|
|
chr7_-_77045706
|
3.093
|
NM_017439
|
PION
|
pigeon homolog (Drosophila)
|
|
chr12_+_107349673
|
2.662
|
|
C12orf23
|
chromosome 12 open reading frame 23
|
|
chr7_-_98741681
|
2.577
|
NM_001199847
NM_020429
NM_181349
|
SMURF1
|
SMAD specific E3 ubiquitin protein ligase 1
|
|
chr3_-_145878775
|
2.557
|
|
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2
|
|
chr2_-_224903933
|
2.499
|
NM_001136528
NM_006216
|
SERPINE2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2
|
|
chr2_-_10588451
|
2.490
|
NM_002539
|
ODC1
|
ornithine decarboxylase 1
|
|
chr4_+_154605421
|
2.415
|
NM_003264
|
TLR2
|
toll-like receptor 2
|
|
chr4_+_154605441
|
2.388
|
|
TLR2
|
toll-like receptor 2
|
|
chr6_-_32805819
|
2.368
|
|
TAP2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP)
|
|
chrX_-_13956530
|
2.323
|
|
GPM6B
|
glycoprotein M6B
|
|
chr1_+_193091283
|
2.322
|
|
CDC73
|
cell division cycle 73, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae)
|
|
chr19_+_14544216
|
2.214
|
|
PKN1
|
protein kinase N1
|
|
chr17_+_79651042
|
2.191
|
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate
|
|
chr12_+_107349539
|
2.148
|
NM_152261
|
C12orf23
|
chromosome 12 open reading frame 23
|
|
chr5_-_150460550
|
2.118
|
|
TNIP1
|
TNFAIP3 interacting protein 1
|
|
chr3_+_23244486
|
2.069
|
|
UBE2E2
|
ubiquitin-conjugating enzyme E2E 2
|
|
chr10_-_133795447
|
2.045
|
|
BNIP3
|
BCL2/adenovirus E1B 19kDa interacting protein 3
|
|
chr10_+_91174438
|
2.039
|
|
IFIT5
|
interferon-induced protein with tetratricopeptide repeats 5
|
|
chr5_-_150460599
|
1.989
|
NM_001252385
NM_001252386
NM_001252393
NM_006058
|
TNIP1
|
TNFAIP3 interacting protein 1
|
|
chr17_+_46985720
|
1.982
|
NM_023079
|
UBE2Z
|
ubiquitin-conjugating enzyme E2Z
|
|
chr10_-_79789290
|
1.974
|
NM_007055
|
POLR3A
|
polymerase (RNA) III (DNA directed) polypeptide A, 155kDa
|
|
chr11_+_65292526
|
1.959
|
NM_001048218
NM_020680
|
SCYL1
|
SCY1-like 1 (S. cerevisiae)
|
|
chr19_+_14544100
|
1.954
|
NM_002741
|
PKN1
|
protein kinase N1
|
|
chr7_+_156931609
|
1.937
|
|
UBE3C
|
ubiquitin protein ligase E3C
|
|
chr18_+_43913906
|
1.927
|
|
RNF165
|
ring finger protein 165
|
|
chr12_-_121734543
|
1.922
|
NM_172226
|
CAMKK2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta
|
|
chr16_-_87903028
|
1.909
|
NM_003486
|
SLC7A5
|
solute carrier family 7 (amino acid transporter light chain, L system), member 5
|
|
chr7_+_156931663
|
1.902
|
|
UBE3C
|
ubiquitin protein ligase E3C
|
|
chr10_-_133795311
|
1.880
|
NM_004052
|
BNIP3
|
BCL2/adenovirus E1B 19kDa interacting protein 3
|
|
chr7_-_27170351
|
1.879
|
NM_002141
|
HOXA4
|
homeobox A4
|
|
chr5_+_17217474
|
1.875
|
NM_006317
|
BASP1
|
brain abundant, membrane attached signal protein 1
|
|
chr6_+_71998514
|
1.864
|
|
OGFRL1
|
opioid growth factor receptor-like 1
|
|
chr5_+_149865310
|
1.843
|
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1
|
|
chr17_-_36831156
|
1.839
|
NM_001130677
|
C17orf96
|
chromosome 17 open reading frame 96
|
|
chr15_+_67358194
|
1.837
|
NM_005902
|
SMAD3
|
SMAD family member 3
|
|
chr17_+_46985829
|
1.833
|
|
UBE2Z
|
ubiquitin-conjugating enzyme E2Z
|
|
chr11_+_65292576
|
1.824
|
|
SCYL1
|
SCY1-like 1 (S. cerevisiae)
|
|
chr13_+_115079943
|
1.821
|
NM_001164144
NM_001164145
NM_032436
|
CHAMP1
|
chromosome alignment maintaining phosphoprotein 1
|
|
chr17_+_46985853
|
1.817
|
|
UBE2Z
|
ubiquitin-conjugating enzyme E2Z
|
|
chr21_+_42797977
|
1.805
|
NM_002462
NM_001178046
|
MX1
|
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse)
|
|
chr5_+_17217781
|
1.788
|
|
BASP1
|
brain abundant, membrane attached signal protein 1
|
|
chr7_+_128095881
|
1.773
|
NM_001098786
NM_013332
|
HILPDA
|
hypoxia inducible lipid droplet-associated
|
|
chr21_+_45719921
|
1.738
|
NM_002626
|
PFKL
|
phosphofructokinase, liver
|
|
chr1_+_227058241
|
1.730
|
NM_000447
NM_012486
|
PSEN2
|
presenilin 2 (Alzheimer disease 4)
|
|
chr6_+_34204928
|
1.728
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr12_-_77459198
|
1.724
|
NM_203394
|
E2F7
|
E2F transcription factor 7
|
|
chr6_+_34204690
|
1.707
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr9_+_35732498
|
1.701
|
|
CREB3
|
cAMP responsive element binding protein 3
|
|
chr1_-_212873104
|
1.698
|
|
BATF3
|
basic leucine zipper transcription factor, ATF-like 3
|
|
chr3_+_23244703
|
1.697
|
NM_152653
|
UBE2E2
|
ubiquitin-conjugating enzyme E2E 2
|
|
chr1_-_38218573
|
1.694
|
|
EPHA10
|
EPH receptor A10
|
|
chr21_+_45719964
|
1.682
|
|
PFKL
|
phosphofructokinase, liver
|
|
chr12_-_121734474
|
1.669
|
|
CAMKK2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta
|
|
chr15_+_67358161
|
1.659
|
|
SMAD3
|
SMAD family member 3
|
|
chr17_+_79651072
|
1.644
|
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate
|
|
chr22_-_50964573
|
1.621
|
NM_001169110
|
SCO2
|
SCO cytochrome oxidase deficient homolog 2 (yeast)
|
|
chr19_-_5720136
|
1.619
|
|
LONP1
|
lon peptidase 1, mitochondrial
|
|
chrX_-_152939256
|
1.609
|
|
PNCK
|
pregnancy up-regulated non-ubiquitously expressed CaM kinase
|
|
chr21_+_26934440
|
1.599
|
|
MIR155HG
|
MIR155 host gene (non-protein coding)
|
|
chr16_+_30709913
|
1.595
|
|
|
|
|
chr6_+_71998476
|
1.573
|
NM_024576
|
OGFRL1
|
opioid growth factor receptor-like 1
|
|
chr8_-_145515014
|
1.560
|
|
BOP1
|
block of proliferation 1
|
|
chr22_-_50964856
|
1.550
|
NM_001169109
|
SCO2
|
SCO cytochrome oxidase deficient homolog 2 (yeast)
|
|
chr6_-_32806009
|
1.521
|
|
TAP2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP)
|
|
chr1_+_193091165
|
1.518
|
|
CDC73
|
cell division cycle 73, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae)
|
|
chr1_+_233749749
|
1.510
|
NM_002245
|
KCNK1
|
potassium channel, subfamily K, member 1
|
|
chr10_+_111767877
|
1.498
|
|
ADD3
|
adducin 3 (gamma)
|
|
chr17_-_41622727
|
1.487
|
|
ETV4
|
ets variant 4
|
|
chr1_-_6662681
|
1.483
|
|
KLHL21
|
kelch-like 21 (Drosophila)
|
|
chr1_+_193091220
|
1.481
|
|
CDC73
|
cell division cycle 73, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae)
|
|
chr7_-_97601654
|
1.480
|
|
|
|
|
chr8_-_30515665
|
1.476
|
NM_002095
|
GTF2E2
|
general transcription factor IIE, polypeptide 2, beta 34kDa
|
|
chr7_-_98741605
|
1.455
|
|
SMURF1
|
SMAD specific E3 ubiquitin protein ligase 1
|
|
chr18_+_55102777
|
1.455
|
NM_004852
|
ONECUT2
|
one cut homeobox 2
|
|
chr1_-_8939150
|
1.450
|
NM_001428
|
ENO1
|
enolase 1, (alpha)
|
|
chr10_-_135171512
|
1.431
|
NM_001098483
NM_198472
|
C10orf125
|
chromosome 10 open reading frame 125
|
|
chr11_+_57435556
|
1.430
|
|
ZDHHC5
|
zinc finger, DHHC-type containing 5
|
|
chr19_-_5720155
|
1.426
|
|
LONP1
|
lon peptidase 1, mitochondrial
|
|
chrX_+_152599544
|
1.418
|
NM_001080485
|
ZNF275
|
zinc finger protein 275
|
|
chr5_-_121413967
|
1.408
|
|
LOX
|
lysyl oxidase
|
|
chr19_+_57154526
|
1.406
|
NM_001193628
|
LOC147670
|
uncharacterized LOC147670
|
|
chr19_-_16683188
|
1.400
|
NM_024881
|
SLC35E1
|
solute carrier family 35, member E1
|
|
chr7_+_156931631
|
1.394
|
NM_014671
|
UBE3C
|
ubiquitin protein ligase E3C
|
|
chr6_+_30539169
|
1.388
|
NM_001025091
NM_001090
|
ABCF1
|
ATP-binding cassette, sub-family F (GCN20), member 1
|
|
chr3_-_150480917
|
1.387
|
|
SIAH2
|
seven in absentia homolog 2 (Drosophila)
|
|
chr7_-_22396511
|
1.382
|
NM_012294
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5
|
|
chr12_+_7023490
|
1.373
|
NM_001975
|
ENO2
|
enolase 2 (gamma, neuronal)
|
|
chr19_+_984316
|
1.367
|
NM_024100
|
WDR18
|
WD repeat domain 18
|
|
chr1_-_111746917
|
1.361
|
|
DENND2D
|
DENN/MADD domain containing 2D
|
|
chr5_-_121413906
|
1.356
|
|
LOX
|
lysyl oxidase
|
|
chr19_-_5720158
|
1.356
|
NM_004793
|
LONP1
|
lon peptidase 1, mitochondrial
|
|
chr3_-_45267804
|
1.355
|
NM_015444
|
TMEM158
|
transmembrane protein 158 (gene/pseudogene)
|
|
chr18_-_71959109
|
1.351
|
NM_001190807
NM_001914
NM_148923
|
CYB5A
|
cytochrome b5 type A (microsomal)
|
|
chr2_-_233352530
|
1.350
|
NM_004826
|
ECEL1
|
endothelin converting enzyme-like 1
|
|
chr3_-_16555128
|
1.346
|
NM_015150
|
RFTN1
|
raftlin, lipid raft linker 1
|
|
chr1_-_41328002
|
1.341
|
NM_133467
|
CITED4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4
|
|
chr1_-_148176392
|
1.341
|
|
|
|
|
chr6_+_71998508
|
1.340
|
|
OGFRL1
|
opioid growth factor receptor-like 1
|
|
chr7_-_108096693
|
1.332
|
NM_001193582
NM_001193583
NM_001193584
NM_005010
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr6_-_101329197
|
1.331
|
NM_006828
NM_022091
|
ASCC3
|
activating signal cointegrator 1 complex subunit 3
|
|
chr1_+_193091196
|
1.330
|
|
CDC73
|
cell division cycle 73, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae)
|
|
chr3_-_52090090
|
1.321
|
NM_001947
|
DUSP7
|
dual specificity phosphatase 7
|
|
chr3_-_150480963
|
1.319
|
NM_005067
|
SIAH2
|
seven in absentia homolog 2 (Drosophila)
|
|
chr3_+_113666544
|
1.319
|
NM_173570
|
ZDHHC23
|
zinc finger, DHHC-type containing 23
|
|
chr9_-_136857285
|
1.312
|
NM_001134398
NM_003371
|
VAV2
|
vav 2 guanine nucleotide exchange factor
|
|
chr2_-_10588273
|
1.311
|
|
ODC1
|
ornithine decarboxylase 1
|
|
chr16_-_75590107
|
1.302
|
NM_001077416
NM_001077418
NM_001077419
|
TMEM231
|
transmembrane protein 231
|
|
chr17_-_41623246
|
1.301
|
NM_001986
|
ETV4
|
ets variant 4
|
|
chr3_-_150480887
|
1.295
|
|
SIAH2
|
seven in absentia homolog 2 (Drosophila)
|
|
chr5_+_98104998
|
1.291
|
NM_001012761
|
RGMB
|
RGM domain family, member B
|
|
chr11_+_57435458
|
1.290
|
NM_015457
|
ZDHHC5
|
zinc finger, DHHC-type containing 5
|
|
chr6_-_101329006
|
1.284
|
|
ASCC3
|
activating signal cointegrator 1 complex subunit 3
|
|
chr15_-_83953465
|
1.266
|
NM_001717
|
BNC1
|
basonuclin 1
|
|
chr11_-_568419
|
1.258
|
|
MIR210HG
|
MIR210 host gene (non-protein coding)
|
|
chr8_-_125740650
|
1.246
|
NM_014751
|
MTSS1
|
metastasis suppressor 1
|
|
chr4_-_120548329
|
1.233
|
NM_033430
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific
|
|
chr4_+_38869346
|
1.224
|
NM_138389
|
FAM114A1
|
family with sequence similarity 114, member A1
|
|
chr6_+_34205369
|
1.223
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr4_+_146540522
|
1.222
|
NM_172250
|
MMAA
|
methylmalonic aciduria (cobalamin deficiency) cblA type
|
|
chr14_-_24740780
|
1.219
|
|
RABGGTA
|
Rab geranylgeranyltransferase, alpha subunit
|
|
chr17_+_45000483
|
1.208
|
NM_001012511
NM_004287
NM_054022
|
GOSR2
|
golgi SNAP receptor complex member 2
|
|
chr13_+_41363754
|
1.194
|
|
SLC25A15
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15
|
|
chr5_+_127419407
|
1.191
|
NM_001046
|
SLC12A2
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 2
|
|
chr15_+_98503907
|
1.191
|
NM_183376
|
ARRDC4
|
arrestin domain containing 4
|
|
chr15_+_98503938
|
1.179
|
|
ARRDC4
|
arrestin domain containing 4
|
|
chr7_-_102715171
|
1.177
|
NM_001111038
|
FBXL13
|
F-box and leucine-rich repeat protein 13
|
|
chr3_-_50374795
|
1.167
|
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1
|
|
chr20_-_590909
|
1.163
|
NM_004609
|
TCF15
|
transcription factor 15 (basic helix-loop-helix)
|
|
chr2_-_113012601
|
1.158
|
NM_032494
|
ZC3H8
|
zinc finger CCCH-type containing 8
|
|
chr21_-_44495894
|
1.158
|
NM_000071
|
CBS
|
cystathionine-beta-synthase
|
|
chr11_+_57435311
|
1.142
|
|
ZDHHC5
|
zinc finger, DHHC-type containing 5
|
|
chr5_-_121414011
|
1.135
|
NM_002317
|
LOX
|
lysyl oxidase
|
|
chr16_+_31191430
|
1.135
|
NM_001170634
NM_001170937
NM_004960
|
FUS
|
fused in sarcoma
|
|
chr2_-_214016332
|
1.134
|
NM_001079526
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr2_-_75062214
|
1.122
|
|
|
|
|
chr1_+_1243971
|
1.119
|
NM_153339
|
PUSL1
|
pseudouridylate synthase-like 1
|
|
chr2_+_65216526
|
1.115
|
|
SLC1A4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4
|
|
chr3_+_133292557
|
1.112
|
|
CDV3
|
CDV3 homolog (mouse)
|
|
chr17_+_45000493
|
1.112
|
|
GOSR2
|
golgi SNAP receptor complex member 2
|
|
chr1_+_166808721
|
1.103
|
NM_017542
|
POGK
|
pogo transposable element with KRAB domain
|
|
chr6_+_33589045
|
1.096
|
NM_002224
|
ITPR3
|
inositol 1,4,5-trisphosphate receptor, type 3
|
|
chr19_+_50887592
|
1.096
|
NM_002691
|
POLD1
|
polymerase (DNA directed), delta 1, catalytic subunit 125kDa
|
|
chr10_+_102747291
|
1.094
|
NM_001163812
NM_001163813
NM_001163814
NM_021830
|
C10orf2
|
chromosome 10 open reading frame 2
|
|
chrX_-_40506800
|
1.092
|
NM_144970
|
CXorf38
|
chromosome X open reading frame 38
|
|
chr14_-_53162418
|
1.091
|
NM_014584
|
ERO1L
|
ERO1-like (S. cerevisiae)
|
|
chr2_-_213403280
|
1.088
|
NM_001042599
NM_005235
|
ERBB4
|
v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian)
|
|
chr1_+_117452359
|
1.087
|
NM_020440
|
PTGFRN
|
prostaglandin F2 receptor negative regulator
|
|
chr11_+_65029312
|
1.083
|
NM_002689
|
POLA2
|
polymerase (DNA directed), alpha 2 (70kD subunit)
|
|
chr20_-_3996035
|
1.080
|
NM_007219
NM_001134337
NM_001134338
|
RNF24
|
ring finger protein 24
|
|
chr10_+_101419169
|
1.078
|
NM_020354
|
ENTPD7
|
ectonucleoside triphosphate diphosphohydrolase 7
|
|
chr1_-_8938691
|
1.077
|
|
ENO1
|
enolase 1, (alpha)
|
|
chr11_+_33061560
|
1.075
|
NM_001145541
|
TCP11L1
|
t-complex 11 (mouse)-like 1
|
|
chr9_+_35732207
|
1.064
|
NM_006368
|
CREB3
|
cAMP responsive element binding protein 3
|
|
chr5_+_133861685
|
1.063
|
NM_015288
|
PHF15
|
PHD finger protein 15
|
|
chr2_+_173420873
|
1.054
|
|
PDK1
|
pyruvate dehydrogenase kinase, isozyme 1
|
|
chr6_+_34204663
|
1.053
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr3_-_179169180
|
1.052
|
|
GNB4
|
guanine nucleotide binding protein (G protein), beta polypeptide 4
|
|
chr10_+_60936346
|
1.043
|
NM_032439
|
PHYHIPL
|
phytanoyl-CoA 2-hydroxylase interacting protein-like
|
|
chr3_-_16554992
|
1.041
|
|
RFTN1
|
raftlin, lipid raft linker 1
|
|
chr3_-_122746544
|
1.034
|
|
SEMA5B
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5B
|
|
chr6_+_43738757
|
1.034
|
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr21_+_43639202
|
1.034
|
NM_004915
NM_016818
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1
|
|
chr11_+_33061291
|
1.033
|
|
TCP11L1
|
t-complex 11 (mouse)-like 1
|
|
chr19_+_38810540
|
1.026
|
|
KCNK6
|
potassium channel, subfamily K, member 6
|
|
chrX_-_152938742
|
1.007
|
NM_001135740
|
PNCK
|
pregnancy up-regulated non-ubiquitously expressed CaM kinase
|
|
chr1_-_159895279
|
1.003
|
NM_003564
|
TAGLN2
|
transgelin 2
|
|
chr12_-_106641675
|
0.994
|
NM_006825
|
CKAP4
|
cytoskeleton-associated protein 4
|
|
chr11_+_576482
|
0.993
|
NM_020901
|
PHRF1
|
PHD and ring finger domains 1
|
|
chr6_-_31324564
|
0.989
|
|
HLA-B
HLA-C
|
major histocompatibility complex, class I, B
major histocompatibility complex, class I, C
|
|
chr5_+_156693051
|
0.984
|
NM_001037333
NM_001037332
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr3_+_25831548
|
0.973
|
NM_001145391
NM_017897
|
OXSM
|
3-oxoacyl-ACP synthase, mitochondrial
|
|
chr17_+_55162530
|
0.972
|
NM_003488
|
AKAP1
|
A kinase (PRKA) anchor protein 1
|
|
chr3_-_50329702
|
0.971
|
NM_006764
|
IFRD2
|
interferon-related developmental regulator 2
|
|
chr11_-_207403
|
0.970
|
NM_001098787
NM_016526
|
BET1L
|
blocked early in transport 1 homolog (S. cerevisiae)-like
|
|
chr9_-_135545327
|
0.970
|
|
DDX31
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31
|
|
chr1_+_184356364
|
0.968
|
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr14_+_55034633
|
0.967
|
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr3_-_156272909
|
0.966
|
NM_007107
|
SSR3
|
signal sequence receptor, gamma (translocon-associated protein gamma)
|
|
chr14_+_70078309
|
0.965
|
NM_014734
|
KIAA0247
|
KIAA0247
|
|
chr17_-_2614926
|
0.965
|
NM_015229
|
KIAA0664
|
KIAA0664
|
|
chr5_+_55033844
|
0.962
|
NM_001142549
NM_001166533
NM_024415
|
DDX4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4
|
|
chr11_-_59436488
|
0.962
|
NM_152716
|
PATL1
|
protein associated with topoisomerase II homolog 1 (yeast)
|