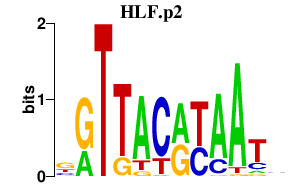
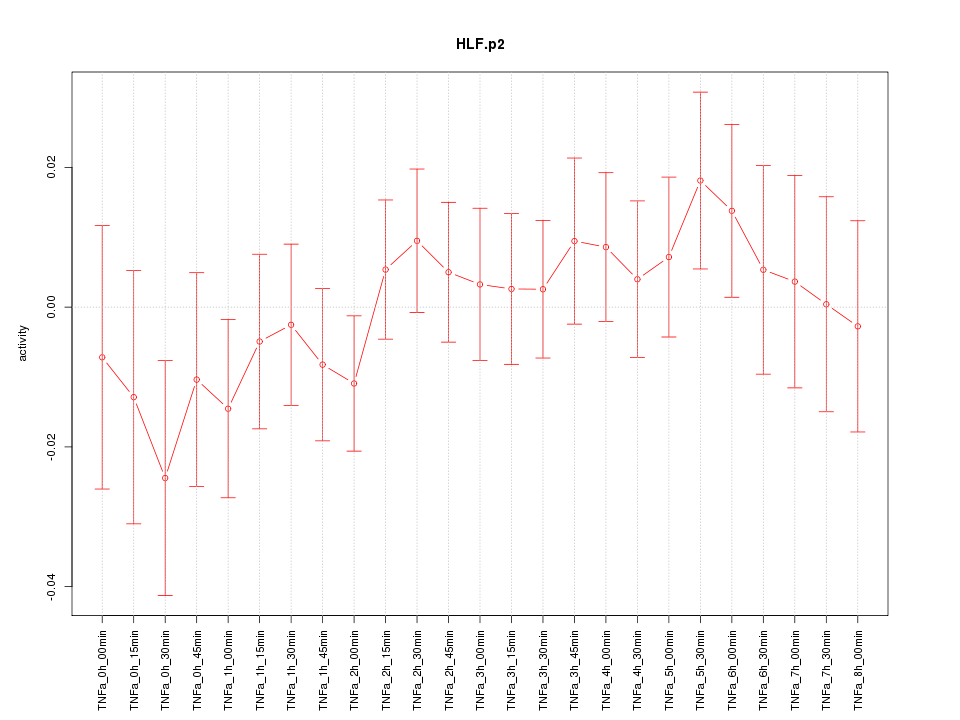
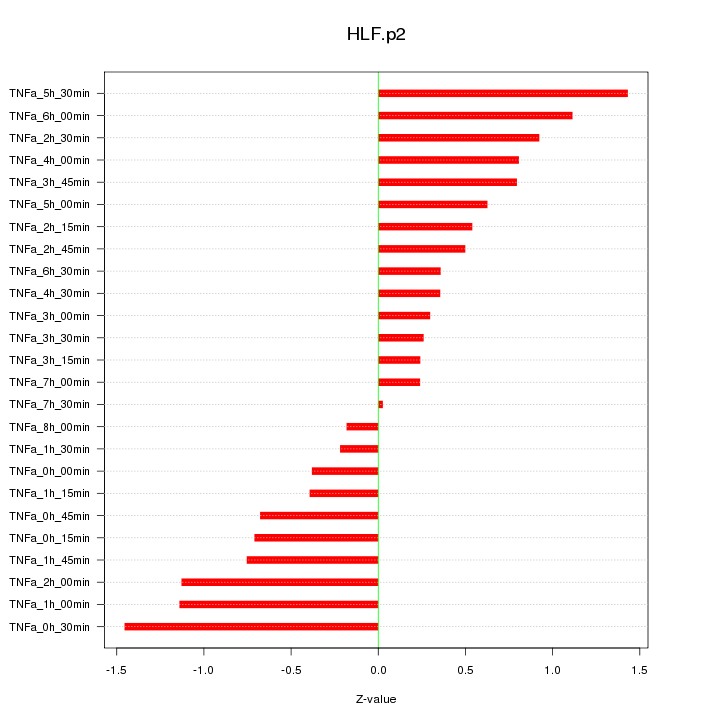
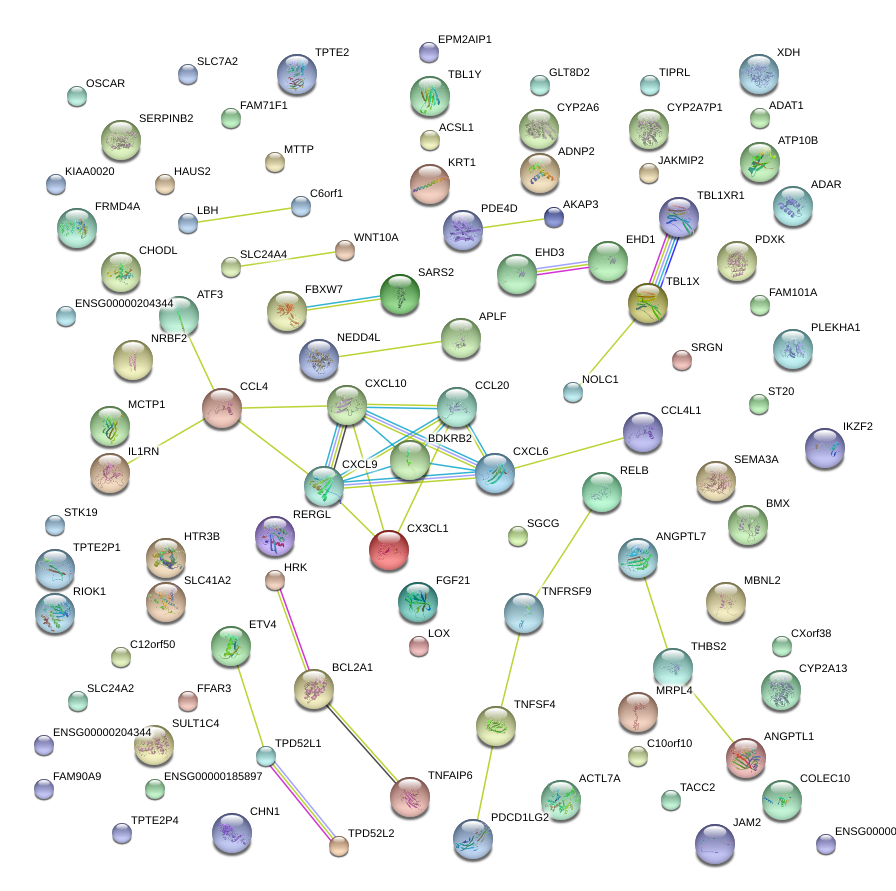
|
chr16_+_57406398
|
4.602
|
NM_002996
|
CX3CL1
|
chemokine (C-X3-C motif) ligand 1
|
|
chr12_-_105322056
|
3.476
|
NM_032148
|
SLC41A2
|
solute carrier family 41, member 2
|
|
chr2_+_152214105
|
2.226
|
NM_007115
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6
|
|
chr19_+_45504686
|
2.007
|
NM_006509
|
RELB
|
v-rel reticuloendotheliosis viral oncogene homolog B
|
|
chr1_-_173174611
|
1.903
|
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4
|
|
chr9_+_111624545
|
1.741
|
NM_006687
|
ACTL7A
|
actin-like 7A
|
|
chr4_+_74702272
|
1.609
|
NM_002993
|
CXCL6
|
chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2)
|
|
chr12_+_124773709
|
1.545
|
NM_181709
|
FAM101A
|
family with sequence similarity 101, member A
|
|
chr11_-_64647136
|
1.491
|
|
EHD1
|
EH-domain containing 1
|
|
chr12_-_117319231
|
1.235
|
NM_003806
|
HRK
|
harakiri, BCL2 interacting protein (contains only BH3 domain)
|
|
chr15_-_80263460
|
1.176
|
NM_001114735
NM_004049
|
BCL2A1
|
BCL2-related protein A1
|
|
chr6_-_169654047
|
1.158
|
NM_003247
|
THBS2
|
thrombospondin 2
|
|
chr5_-_58882274
|
1.148
|
NM_006203
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr18_+_61554938
|
1.129
|
NM_001143818
NM_002575
|
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2
|
|
chr6_-_169653922
|
1.092
|
|
THBS2
|
thrombospondin 2
|
|
chr1_-_8003224
|
1.088
|
NM_001561
|
TNFRSF9
|
tumor necrosis factor receptor superfamily, member 9
|
|
chr10_-_14372807
|
1.006
|
NM_018027
|
FRMD4A
|
FERM domain containing 4A
|
|
chr7_-_83824216
|
0.977
|
NM_006080
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A
|
|
chr17_+_34431219
|
0.971
|
NM_002984
|
CCL4
|
chemokine (C-C motif) ligand 4
|
|
chr13_+_97928414
|
0.963
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr2_+_68694669
|
0.928
|
NM_173545
|
APLF
|
aprataxin and PNKP like factor
|
|
chr10_+_123923104
|
0.905
|
NM_006997
NM_206860
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr1_-_154600383
|
0.846
|
|
ADAR
|
adenosine deaminase, RNA-specific
|
|
chr4_-_76928601
|
0.821
|
NM_002416
|
CXCL9
|
chemokine (C-X-C motif) ligand 9
|
|
chr4_-_76944585
|
0.820
|
NM_001565
|
CXCL10
|
chemokine (C-X-C motif) ligand 10
|
|
chr18_+_77866914
|
0.805
|
NM_014913
|
ADNP2
|
ADNP homeobox 2
|
|
chr1_-_154600446
|
0.734
|
NM_001025107
|
ADAR
|
adenosine deaminase, RNA-specific
|
|
chr19_+_41594367
|
0.719
|
NM_000766
|
CYP2A13
|
cytochrome P450, family 2, subfamily A, polypeptide 13
|
|
chr2_-_175712276
|
0.718
|
NM_001206602
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr19_+_35849487
|
0.716
|
NM_005304
|
FFAR3
|
free fatty acid receptor 3
|
|
chr9_+_5510544
|
0.708
|
NM_025239
|
PDCD1LG2
|
programmed cell death 1 ligand 2
|
|
chr5_-_147162251
|
0.703
|
NM_014790
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr17_-_41623246
|
0.701
|
NM_001986
|
ETV4
|
ets variant 4
|
|
chr21_+_27011588
|
0.694
|
NM_021219
|
JAM2
|
junctional adhesion molecule 2
|
|
chr10_+_124151818
|
0.684
|
NM_021622
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chr1_+_212738675
|
0.671
|
NM_001030287
|
ATF3
|
activating transcription factor 3
|
|
chr2_+_113885137
|
0.663
|
NM_173842
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr19_+_10362886
|
0.659
|
|
MRPL4
|
mitochondrial ribosomal protein L4
|
|
chr6_+_7402879
|
0.654
|
NM_153005
|
RIOK1
|
RIO kinase 1 (yeast)
|
|
chr13_+_23755059
|
0.649
|
NM_000231
|
SGCG
|
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein)
|
|
chr2_+_108994366
|
0.646
|
NM_006588
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4
|
|
chr10_+_70847817
|
0.631
|
NM_002727
|
SRGN
|
serglycin
|
|
chr1_+_168148082
|
0.622
|
NM_001031800
NM_152902
|
TIPRL
|
TIP41, TOR signaling pathway regulator-like (S. cerevisiae)
|
|
chr8_+_17396285
|
0.618
|
NM_001164771
NM_003046
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr2_-_214015053
|
0.617
|
NM_016260
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr4_-_185747183
|
0.617
|
NM_001995
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1
|
|
chr18_+_55888799
|
0.606
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chrX_+_9431314
|
0.584
|
NM_001139466
NM_001139467
NM_001139468
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr10_+_103911932
|
0.577
|
NM_004741
|
NOLC1
|
nucleolar and coiled-body phosphoprotein 1
|
|
chr7_+_45933096
|
0.571
|
|
|
|
|
chr6_+_31940098
|
0.568
|
|
STK19
|
serine/threonine kinase 19
|
|
chr7_+_128349111
|
0.565
|
|
FAM71F1
|
family with sequence similarity 71, member F1
|
|
chr18_+_55888750
|
0.564
|
NM_001144966
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr4_-_153273683
|
0.560
|
NM_018315
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr5_-_121413906
|
0.547
|
|
LOX
|
lysyl oxidase
|
|
chr1_-_178840079
|
0.537
|
NM_004673
|
ANGPTL1
|
angiopoietin-like 1
|
|
chr12_-_4754275
|
0.537
|
NM_006422
|
AKAP3
|
A kinase (PRKA) anchor protein 3
|
|
chr2_+_113885145
|
0.530
|
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr11_+_113775517
|
0.514
|
NM_006028
|
HTR3B
|
5-hydroxytryptamine (serotonin) receptor 3B
|
|
chr19_+_10362912
|
0.510
|
|
MRPL4
|
mitochondrial ribosomal protein L4
|
|
chr9_-_19786886
|
0.506
|
NM_001193288
NM_020344
|
SLC24A2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2
|
|
chr12_-_53074172
|
0.504
|
NM_006121
|
KRT1
|
keratin 1
|
|
chr5_-_121413967
|
0.490
|
|
LOX
|
lysyl oxidase
|
|
chr13_-_20110884
|
0.484
|
NM_001141968
NM_130785
NM_199254
|
TPTE2
|
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2
|
|
chr2_+_228678556
|
0.482
|
NM_001130046
NM_004591
|
CCL20
|
chemokine (C-C motif) ligand 20
|
|
chr4_+_100485239
|
0.480
|
NM_000253
|
MTTP
|
microsomal triglyceride transfer protein
|
|
chr1_-_173176314
|
0.471
|
NM_003326
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4
|
|
chr10_+_70847857
|
0.471
|
|
SRGN
|
serglycin
|
|
chr18_+_55816546
|
0.468
|
NM_001144965
NM_001144968
NM_001144969
NM_001144971
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr19_-_54604070
|
0.468
|
NM_130771
NM_133168
NM_133169
NM_206818
|
OSCAR
|
osteoclast associated, immunoglobulin-like receptor
|
|
chr21_+_19617145
|
0.464
|
NM_024944
|
CHODL
|
chondrolectin
|
|
chr10_-_45474210
|
0.462
|
NM_007021
|
C10orf10
|
chromosome 10 open reading frame 10
|
|
chr1_+_11249344
|
0.460
|
NM_021146
|
ANGPTL7
|
angiopoietin-like 7
|
|
chr4_-_185747051
|
0.457
|
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1
|
|
chr14_+_96671015
|
0.451
|
NM_000623
|
BDKRB2
|
bradykinin receptor B2
|
|
chr9_-_2844098
|
0.448
|
NM_014878
|
KIAA0020
|
KIAA0020
|
|
chrX_+_15518899
|
0.442
|
NM_203281
|
BMX
|
BMX non-receptor tyrosine kinase
|
|
chr16_-_75657153
|
0.439
|
NM_012091
|
ADAT1
|
adenosine deaminase, tRNA-specific 1
|
|
chr2_+_30454396
|
0.434
|
NM_030915
|
LBH
|
limb bud and heart development homolog (mouse)
|
|
chr9_-_2844083
|
0.433
|
|
KIAA0020
|
KIAA0020
|
|
chr7_-_45026204
|
0.430
|
|
SNHG15
|
small nucleolar RNA host gene 15 (non-protein coding)
|
|
chr2_-_31637553
|
0.426
|
|
XDH
|
xanthine dehydrogenase
|
|
chr5_-_160279045
|
0.419
|
NM_025153
|
ATP10B
|
ATPase, class V, type 10B
|
|
chr10_+_64893003
|
0.418
|
NM_030759
|
NRBF2
|
nuclear receptor binding factor 2
|
|
chr21_+_45138987
|
0.417
|
|
PDXK
|
pyridoxal (pyridoxine, vitamin B6) kinase
|
|
chr2_+_219745243
|
0.416
|
NM_025216
|
WNT10A
|
wingless-type MMTV integration site family, member 10A
|
|
chr5_-_94417544
|
0.410
|
NM_001002796
|
MCTP1
|
multiple C2 domains, transmembrane 1
|
|
chr12_-_88423145
|
0.410
|
NM_152589
|
C12orf50
|
chromosome 12 open reading frame 50
|
|
chr15_+_42840991
|
0.404
|
NM_001130447
NM_018097
|
HAUS2
|
HAUS augmin-like complex, subunit 2
|
|
chr8_+_120079423
|
0.401
|
NM_006438
|
COLEC10
|
collectin sub-family member 10 (C-type lectin)
|
|
chr19_+_49259343
|
0.401
|
NM_019113
|
FGF21
|
fibroblast growth factor 21
|
|
chrX_-_40506666
|
0.398
|
|
CXorf38
|
chromosome X open reading frame 38
|
|
chr20_+_62496634
|
0.376
|
|
TPD52L2
|
tumor protein D52-like 2
|
|
chr20_+_62496650
|
0.375
|
|
TPD52L2
|
tumor protein D52-like 2
|
|
chr6_-_13295817
|
0.372
|
NM_001242698
|
LOC100130357
|
uncharacterized LOC100130357
|
|
chr20_+_36011936
|
0.359
|
|
SRC
|
v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian)
|
|
chr21_+_45138973
|
0.354
|
NM_003681
|
PDXK
|
pyridoxal (pyridoxine, vitamin B6) kinase
|
|
chrX_+_110339374
|
0.349
|
NM_002578
|
PAK3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr5_-_147162264
|
0.345
|
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr20_+_62496678
|
0.343
|
|
TPD52L2
|
tumor protein D52-like 2
|
|
chr12_+_112856756
|
0.342
|
|
PTPN11
|
protein tyrosine phosphatase, non-receptor type 11
|
|
chr18_+_55888838
|
0.340
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr1_+_168148215
|
0.339
|
|
TIPRL
|
TIP41, TOR signaling pathway regulator-like (S. cerevisiae)
|
|
chr8_+_124194873
|
0.337
|
|
FAM83A
|
family with sequence similarity 83, member A
|
|
chrX_+_65235279
|
0.337
|
|
|
|
|
chr3_-_99594915
|
0.333
|
NM_014890
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr7_+_28725597
|
0.333
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr10_+_114043413
|
0.332
|
NM_058222
|
TECTB
|
tectorin beta
|
|
chr17_-_55980749
|
0.327
|
NM_017949
|
CUEDC1
|
CUE domain containing 1
|
|
chr9_-_73029539
|
0.326
|
NM_001206
|
KLF9
|
Kruppel-like factor 9
|
|
chr9_-_95055966
|
0.325
|
NM_013417
|
IARS
|
isoleucyl-tRNA synthetase
|
|
chr14_-_77889231
|
0.324
|
NM_001113475
|
NOXRED1
|
NADP-dependent oxidoreductase domain containing 1
|
|
chr7_-_31380507
|
0.320
|
NM_022728
|
NEUROD6
|
neurogenic differentiation 6
|
|
chr5_-_138718941
|
0.318
|
|
SLC23A1
|
solute carrier family 23 (nucleobase transporters), member 1
|
|
chr11_-_14993790
|
0.316
|
NM_001033952
NM_001033953
NM_001741
|
CALCA
|
calcitonin-related polypeptide alpha
|
|
chr3_-_15382874
|
0.312
|
NM_001018009
|
SH3BP5
|
SH3-domain binding protein 5 (BTK-associated)
|
|
chr7_-_112727832
|
0.312
|
NM_001146265
NM_018970
|
GPR85
|
G protein-coupled receptor 85
|
|
chr12_-_123380608
|
0.310
|
|
|
|
|
chr10_+_91152321
|
0.310
|
NM_001548
|
IFIT1
|
interferon-induced protein with tetratricopeptide repeats 1
|
|
chr1_-_21501229
|
0.308
|
NM_001198803
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3
|
|
chr2_-_113594325
|
0.307
|
NM_000576
|
IL1B
|
interleukin 1, beta
|
|
chr6_-_90024936
|
0.307
|
NM_002043
|
GABRR2
|
gamma-aminobutyric acid (GABA) receptor, rho 2
|
|
chr3_-_185655776
|
0.289
|
|
TRA2B
|
transformer 2 beta homolog (Drosophila)
|
|
chr20_+_58296264
|
0.283
|
NM_183244
NM_183246
|
PHACTR3
|
phosphatase and actin regulator 3
|
|
chr15_-_83621350
|
0.279
|
NM_004839
NM_199330
|
HOMER2
|
homer homolog 2 (Drosophila)
|
|
chr9_-_35958095
|
0.278
|
NM_019897
|
OR2S2
|
olfactory receptor, family 2, subfamily S, member 2
|
|
chr21_-_30365111
|
0.277
|
NM_015565
|
LTN1
|
listerin E3 ubiquitin protein ligase 1
|
|
chr9_-_95055986
|
0.277
|
|
IARS
|
isoleucyl-tRNA synthetase
|
|
chr8_+_38585703
|
0.276
|
NM_001146216
|
TACC1
|
transforming, acidic coiled-coil containing protein 1
|
|
chr11_+_5372737
|
0.276
|
NM_001004750
|
OR51B6
|
olfactory receptor, family 51, subfamily B, member 6
|
|
chr19_+_49109999
|
0.276
|
NM_133498
|
SPACA4
|
sperm acrosome associated 4
|
|
chr2_+_191002485
|
0.276
|
NM_001042521
|
C2orf88
|
chromosome 2 open reading frame 88
|
|
chr20_+_55099784
|
0.273
|
NM_001012971
|
FAM209A
FAM209B
|
family with sequence similarity 209, member A
family with sequence similarity 209, member B
|
|
chr1_-_32264271
|
0.272
|
|
SPOCD1
|
SPOC domain containing 1
|
|
chr5_-_94417185
|
0.271
|
|
MCTP1
|
multiple C2 domains, transmembrane 1
|
|
chr1_-_213020947
|
0.271
|
NM_001024601
|
C1orf227
|
chromosome 1 open reading frame 227
|
|
chr8_-_145115565
|
0.267
|
NM_017570
|
OPLAH
|
5-oxoprolinase (ATP-hydrolysing)
|
|
chr11_+_5710926
|
0.266
|
|
TRIM22
|
tripartite motif containing 22
|
|
chr13_-_25746304
|
0.266
|
|
FAM123A
|
family with sequence similarity 123A
|
|
chr7_+_90032647
|
0.264
|
NM_001185072
NM_001185073
NM_012129
|
CLDN12
|
claudin 12
|
|
chr6_-_43595079
|
0.263
|
|
GTPBP2
|
GTP binding protein 2
|
|
chr5_-_138719018
|
0.262
|
NM_005847
NM_152685
|
SLC23A1
|
solute carrier family 23 (nucleobase transporters), member 1
|
|
chr3_-_138478184
|
0.262
|
NM_006219
|
PIK3CB
|
phosphoinositide-3-kinase, catalytic, beta polypeptide
|
|
chr10_+_86004808
|
0.261
|
NM_001012720
NM_001012722
NM_002921
|
RGR
|
retinal G protein coupled receptor
|
|
chr3_-_16306283
|
0.261
|
NM_001047434
NM_206831
|
DPH3
|
DPH3, KTI11 homolog (S. cerevisiae)
|
|
chr8_+_24298442
|
0.260
|
NM_003817
|
ADAM7
|
ADAM metallopeptidase domain 7
|
|
chr17_-_49124128
|
0.257
|
NM_001251971
|
SPAG9
|
sperm associated antigen 9
|
|
chr17_-_34329083
|
0.256
|
NM_032965
|
CCL15
|
chemokine (C-C motif) ligand 15
|
|
chr5_-_32388595
|
0.255
|
|
ZFR
|
zinc finger RNA binding protein
|
|
chr17_+_51900238
|
0.255
|
NM_032559
|
KIF2B
|
kinesin family member 2B
|
|
chr12_-_54758228
|
0.254
|
NM_020370
|
GPR84
|
G protein-coupled receptor 84
|
|
chr16_+_20911987
|
0.250
|
NM_001128301
NM_001128302
|
LYRM1
|
LYR motif containing 1
|
|
chr18_+_77867145
|
0.249
|
|
ADNP2
|
ADNP homeobox 2
|
|
chr22_-_43355886
|
0.249
|
NM_001184971
|
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2
|
|
chr17_+_74536114
|
0.249
|
NM_001077620
|
PRCD
|
progressive rod-cone degeneration
|
|
chr5_+_95998170
|
0.248
|
|
CAST
|
calpastatin
|
|
chr19_+_36119958
|
0.247
|
NM_024321
|
RBM42
|
RNA binding motif protein 42
|
|
chr7_-_115670774
|
0.246
|
|
TFEC
|
transcription factor EC
|
|
chr12_+_57623355
|
0.240
|
NM_001166356
NM_005412
|
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial)
|
|
chr14_+_21249209
|
0.240
|
NM_005615
|
RNASE6
|
ribonuclease, RNase A family, k6
|
|
chr14_-_63785592
|
0.240
|
NM_145171
|
GPHB5
|
glycoprotein hormone beta 5
|
|
chr10_+_118305427
|
0.239
|
NM_000936
|
PNLIP
|
pancreatic lipase
|
|
chr6_-_114384008
|
0.236
|
NM_153612
|
HS3ST5
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 5
|
|
chr17_-_17485691
|
0.235
|
|
PEMT
|
phosphatidylethanolamine N-methyltransferase
|
|
chr19_+_49622480
|
0.233
|
NM_003660
|
PPFIA3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3
|
|
chr10_+_114133775
|
0.232
|
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr5_-_39364562
|
0.232
|
NM_001737
|
C9
|
complement component 9
|
|
chr9_+_36169384
|
0.230
|
NM_005893
|
CCIN
|
calicin
|
|
chr7_-_112727778
|
0.229
|
|
GPR85
|
G protein-coupled receptor 85
|
|
chr12_-_16757944
|
0.229
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr22_-_30867944
|
0.228
|
NM_174975
|
SEC14L3
|
SEC14-like 3 (S. cerevisiae)
|
|
chr20_-_44420538
|
0.228
|
NM_080614
|
WFDC3
|
WAP four-disulfide core domain 3
|
|
chr12_-_75603510
|
0.227
|
NM_139136
NM_139137
NM_153748
|
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2
|
|
chr21_+_45209398
|
0.226
|
NM_003683
|
RRP1
|
ribosomal RNA processing 1 homolog (S. cerevisiae)
|
|
chr8_-_101315457
|
0.226
|
NM_015435
|
RNF19A
|
ring finger protein 19A
|
|
chrX_-_40506800
|
0.225
|
NM_144970
|
CXorf38
|
chromosome X open reading frame 38
|
|
chr13_-_25745620
|
0.225
|
|
FAM123A
|
family with sequence similarity 123A
|
|
chr2_-_73869493
|
0.224
|
NM_003960
|
NAT8
|
N-acetyltransferase 8 (GCN5-related, putative)
|
|
chr20_+_44420606
|
0.223
|
|
DNTTIP1
|
deoxynucleotidyltransferase, terminal, interacting protein 1
|
|
chr17_+_685512
|
0.220
|
NM_018146
|
RNMTL1
|
RNA methyltransferase like 1
|
|
chr3_-_195306274
|
0.219
|
|
APOD
|
apolipoprotein D
|
|
chr7_-_112727799
|
0.218
|
|
GPR85
|
G protein-coupled receptor 85
|
|
chr11_-_77184549
|
0.218
|
NM_001128620
NM_002576
|
PAK1
|
p21 protein (Cdc42/Rac)-activated kinase 1
|
|
chr1_-_93342706
|
0.216
|
NM_001252271
|
FAM69A
|
family with sequence similarity 69, member A
|
|
chr9_-_114247024
|
0.215
|
NM_001080398
|
KIAA0368
|
KIAA0368
|
|
chr11_+_69061599
|
0.215
|
NM_138768
|
MYEOV
|
myeloma overexpressed (in a subset of t(11;14) positive multiple myelomas)
|
|
chr1_+_209878135
|
0.215
|
NM_005525
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1
|
|
chr17_-_39324423
|
0.213
|
NM_033187
|
KRTAP4-3
|
keratin associated protein 4-3
|
|
chr1_-_102462789
|
0.213
|
NM_058170
|
OLFM3
|
olfactomedin 3
|
|
chr2_-_114514196
|
0.213
|
NM_025181
|
SLC35F5
|
solute carrier family 35, member F5
|
|
chr1_-_100643770
|
0.213
|
NM_144620
|
LRRC39
|
leucine rich repeat containing 39
|
|
chr19_+_55014012
|
0.212
|
NM_002288
NM_021270
|
LAIR2
|
leukocyte-associated immunoglobulin-like receptor 2
|
|
chr9_-_94186140
|
0.208
|
NM_005384
|
NFIL3
|
nuclear factor, interleukin 3 regulated
|
|
chr21_+_35736322
|
0.208
|
NM_172201
|
KCNE2
|
potassium voltage-gated channel, Isk-related family, member 2
|
|
chr2_+_162272892
|
0.207
|
|
TBR1
|
T-box, brain, 1
|
|
chr6_+_131894343
|
0.205
|
NM_000045
NM_001244438
|
ARG1
|
arginase, liver
|
|
chr19_-_35992798
|
0.205
|
NM_001035516
|
DMKN
|
dermokine
|
|
chr12_+_57623827
|
0.205
|
NM_001166358
NM_001166359
NM_001166357
|
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial)
|
|
chr13_-_25745778
|
0.202
|
NM_152704
NM_199138
|
FAM123A
|
family with sequence similarity 123A
|
|
chr5_+_6746100
|
0.200
|
NM_001171806
|
PAPD7
|
PAP associated domain containing 7
|