|
chr13_-_72440557
|
4.873
|
|
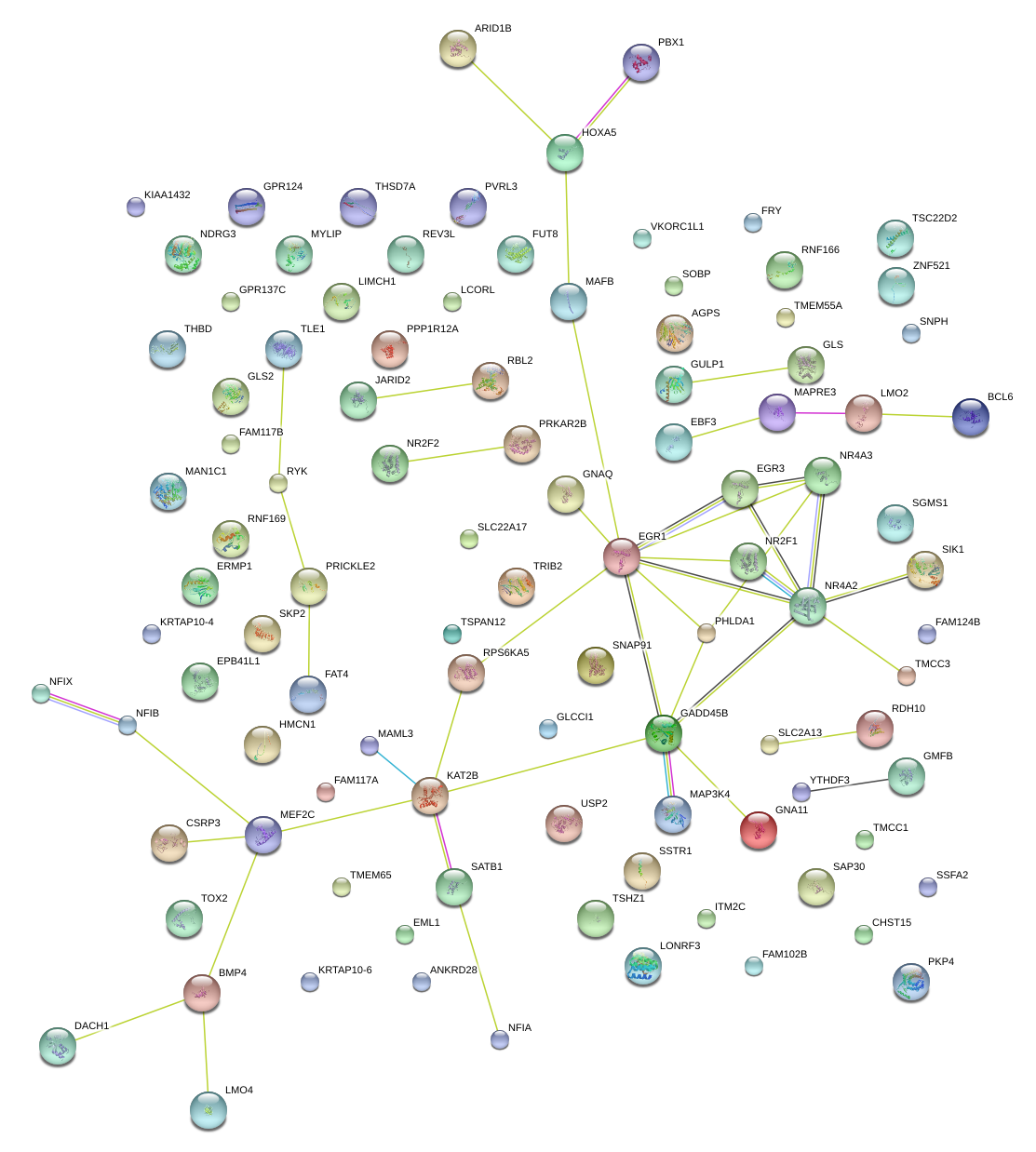
DACH1
|
dachshund homolog 1 (Drosophila)
|
|
chr8_-_22550708
|
4.628
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr7_-_27183225
|
4.519
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr10_-_131762017
|
4.358
|
NM_001005463
|
EBF3
|
early B-cell factor 3
|
|
chr18_-_22931970
|
4.163
|
|
ZNF521
|
zinc finger protein 521
|
|
chr18_-_22932109
|
3.651
|
NM_015461
|
ZNF521
|
zinc finger protein 521
|
|
chr2_+_191745546
|
3.311
|
NM_014905
|
GLS
|
glutaminase
|
|
chr12_-_124068864
|
3.207
|
|
|
|
|
chr17_-_47841468
|
3.128
|
|
FAM117A
|
family with sequence similarity 117, member A
|
|
chr2_-_157189040
|
3.067
|
NM_006186
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chr13_-_72440143
|
3.034
|
|
DACH1
|
dachshund homolog 1 (Drosophila)
|
|
chr3_+_110790460
|
3.003
|
NM_001243286
NM_015480
|
PVRL3
|
poliovirus receptor-related 3
|
|
chr17_-_47841517
|
2.976
|
NM_030802
|
FAM117A
|
family with sequence similarity 117, member A
|
|
chr3_-_64211111
|
2.974
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr4_-_18023358
|
2.880
|
NM_001166139
NM_153686
|
LCORL
|
ligand dependent nuclear receptor corepressor-like
|
|
chr8_-_22550057
|
2.796
|
NM_001199881
|
EGR3
|
early growth response 3
|
|
chr2_+_191745568
|
2.790
|
|
GLS
|
glutaminase
|
|
chr19_+_2476122
|
2.783
|
NM_015675
|
GADD45B
|
growth arrest and DNA-damage-inducible, beta
|
|
chr9_-_80646150
|
2.743
|
NM_002072
|
GNAQ
|
guanine nucleotide binding protein (G protein), q polypeptide
|
|
chr9_-_14313509
|
2.400
|
|
NFIB
|
nuclear factor I/B
|
|
chr20_-_35374530
|
2.309
|
NM_022477
NM_032013
|
NDRG3
|
NDRG family member 3
|
|
chr20_-_39317875
|
2.275
|
NM_005461
|
MAFB
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian)
|
|
chr13_+_32605436
|
2.232
|
NM_023037
|
FRY
|
furry homolog (Drosophila)
|
|
chr14_+_38678280
|
2.222
|
|
SSTR1
|
somatostatin receptor 1
|
|
chr2_+_191745791
|
2.220
|
|
GLS
|
glutaminase
|
|
chr14_-_54423448
|
2.207
|
NM_001202
NM_130850
|
BMP4
|
bone morphogenetic protein 4
|
|
chr12_-_76425207
|
2.202
|
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr2_+_203499819
|
2.185
|
NM_173511
|
FAM117B
|
family with sequence similarity 117, member B
|
|
chr8_-_125384916
|
2.122
|
NM_194291
|
TMEM65
|
transmembrane protein 65
|
|
chr2_+_191745547
|
2.096
|
|
GLS
|
glutaminase
|
|
chr2_-_225266710
|
2.050
|
NM_001122779
NM_024785
|
FAM124B
|
family with sequence similarity 124B
|
|
chr20_-_23030141
|
2.040
|
NM_000361
|
THBD
|
thrombomodulin
|
|
chr2_+_191745912
|
2.013
|
|
GLS
|
glutaminase
|
|
chr4_+_41614725
|
1.987
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr12_-_95044205
|
1.985
|
NM_020698
|
TMCC3
|
transmembrane and coiled-coil domain family 3
|
|
chr11_+_74459900
|
1.946
|
NM_001098638
|
RNF169
|
ring finger protein 169
|
|
chr2_+_189156603
|
1.882
|
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1
|
|
chr1_+_25943958
|
1.829
|
NM_020379
|
MAN1C1
|
mannosidase, alpha, class 1C, member 1
|
|
chr7_+_106685007
|
1.827
|
NM_002736
|
PRKAR2B
|
protein kinase, cAMP-dependent, regulatory, type II, beta
|
|
chr3_+_150125983
|
1.813
|
|
TSC22D2
|
TSC22 domain family, member 2
|
|
chr14_-_91526912
|
1.810
|
NM_004755
NM_182398
|
RPS6KA5
|
ribosomal protein S6 kinase, 90kDa, polypeptide 5
|
|
chr3_+_20081853
|
1.808
|
|
KAT2B
|
K(lysine) acetyltransferase 2B
|
|
chr1_+_87794576
|
1.788
|
|
LMO4
|
LIM domain only 4
|
|
chr4_-_141075232
|
1.753
|
NM_018717
|
MAML3
|
mastermind-like 3 (Drosophila)
|
|
chr6_+_161412807
|
1.715
|
NM_005922
NM_006724
|
MAP3K4
|
mitogen-activated protein kinase kinase kinase 4
|
|
chr15_+_96873845
|
1.705
|
NM_021005
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr20_-_35374468
|
1.702
|
|
NDRG3
|
NDRG family member 3
|
|
chr2_+_12856813
|
1.688
|
NM_021643
|
TRIB2
|
tribbles homolog 2 (Drosophila)
|
|
chr2_+_159313466
|
1.684
|
NM_001005476
NM_003628
|
PKP4
|
plakophilin 4
|
|
chr7_-_120498009
|
1.665
|
|
TSPAN12
|
tetraspanin 12
|
|
chr21_-_46012383
|
1.659
|
NM_198688
|
KRTAP10-6
|
keratin associated protein 10-6
|
|
chr3_-_18466759
|
1.639
|
NM_001195470
NM_002971
|
SATB1
|
SATB homeobox 1
|
|
chr7_+_8008416
|
1.624
|
NM_138426
|
GLCCI1
|
glucocorticoid induced transcript 1
|
|
chr14_-_54955714
|
1.615
|
NM_004124
|
GMFB
|
glia maturation factor, beta
|
|
chr16_+_53468406
|
1.614
|
|
RBL2
|
retinoblastoma-like 2 (p130)
|
|
chr8_-_92053043
|
1.601
|
NM_018710
|
TMEM55A
|
transmembrane protein 55A
|
|
chr6_-_111804413
|
1.577
|
NM_002912
|
REV3L
|
REV3-like, catalytic subunit of DNA polymerase zeta (yeast)
|
|
chr2_+_159313407
|
1.551
|
|
PKP4
|
plakophilin 4
|
|
chr6_+_107811272
|
1.524
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr21_-_44846927
|
1.503
|
NM_173354
|
SIK1
|
salt-inducible kinase 1
|
|
chr10_-_125853102
|
1.493
|
|
CHST15
|
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15
|
|
chr1_+_87794548
|
1.492
|
|
LMO4
|
LIM domain only 4
|
|
chr6_+_15246299
|
1.484
|
NM_004973
|
JARID2
|
jumonji, AT rich interactive domain 2
|
|
chr18_+_77659446
|
1.482
|
|
|
|
|
chr5_+_137801178
|
1.451
|
NM_001964
|
EGR1
|
early growth response 1
|
|
chr20_+_34742656
|
1.389
|
NM_012156
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr16_+_53468373
|
1.385
|
|
RBL2
|
retinoblastoma-like 2 (p130)
|
|
chr4_+_174292100
|
1.373
|
|
SAP30
|
Sin3A-associated protein, 30kDa
|
|
chr20_+_42574535
|
1.352
|
NM_001098798
|
TOX2
|
TOX high mobility group box family member 2
|
|
chr14_-_54955664
|
1.342
|
|
GMFB
|
glia maturation factor, beta
|
|
chr3_-_187463227
|
1.335
|
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr3_-_15900928
|
1.328
|
NM_015199
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr11_-_33891258
|
1.322
|
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr11_-_33891485
|
1.321
|
NM_001142315
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr7_-_120498128
|
1.308
|
NM_012338
|
TSPAN12
|
tetraspanin 12
|
|
chr1_+_109102970
|
1.303
|
NM_001010883
|
FAM102B
|
family with sequence similarity 102, member B
|
|
chr11_-_119252267
|
1.302
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr5_-_88199868
|
1.301
|
NM_001131005
NM_001193347
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr8_-_22549901
|
1.291
|
NM_001199880
|
EGR3
|
early growth response 3
|
|
chr3_-_129612371
|
1.278
|
NM_001128224
|
TMCC1
|
transmembrane and coiled-coil domain family 1
|
|
chr6_+_157099873
|
1.257
|
|
ARID1B
|
AT rich interactive domain 1B (SWI1-like)
|
|
chr4_+_174292027
|
1.256
|
NM_003864
|
SAP30
|
Sin3A-associated protein, 30kDa
|
|
chr9_-_5833072
|
1.248
|
NM_024896
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1
|
|
chr14_+_100259445
|
1.244
|
NM_001008707
NM_004434
|
EML1
|
echinoderm microtubule associated protein like 1
|
|
chr8_+_64081187
|
1.211
|
|
YTHDF3
|
YTH domain family, member 3
|
|
chr20_+_1246959
|
1.209
|
NM_014723
|
SNPH
|
syntaphilin
|
|
chr5_+_36152142
|
1.209
|
NM_001243120
NM_005983
NM_032637
|
SKP2
|
S-phase kinase-associated protein 2 (p45)
|
|
chr4_+_126237566
|
1.209
|
NM_024582
|
FAT4
|
FAT tumor suppressor homolog 4 (Drosophila)
|
|
chr1_+_164528586
|
1.208
|
NM_001204961
NM_001204963
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr8_+_64081108
|
1.191
|
NM_152758
|
YTHDF3
|
YTH domain family, member 3
|
|
chr1_+_87794150
|
1.185
|
NM_006769
|
LMO4
|
LIM domain only 4
|
|
chr16_+_53468390
|
1.180
|
|
RBL2
|
retinoblastoma-like 2 (p130)
|
|
chr8_+_37654757
|
1.176
|
|
GPR124
|
G protein-coupled receptor 124
|
|
chr14_+_65879694
|
1.163
|
|
FUT8
|
fucosyltransferase 8 (alpha (1,6) fucosyltransferase)
|
|
chr12_-_80328684
|
1.160
|
NM_001244992
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|
|
chr6_+_161412685
|
1.156
|
|
MAP3K4
|
mitogen-activated protein kinase kinase kinase 4
|
|
chr8_+_37654406
|
1.156
|
|
GPR124
|
G protein-coupled receptor 124
|
|
chr2_+_189156395
|
1.147
|
NM_001252668
NM_001252669
NM_016315
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1
|
|
chr12_-_40499604
|
1.146
|
NM_052885
|
SLC2A13
|
solute carrier family 2 (facilitated glucose transporter), member 13
|
|
chr7_-_11871735
|
1.141
|
NM_015204
|
THSD7A
|
thrombospondin, type I, domain containing 7A
|
|
chr14_-_23821423
|
1.140
|
NM_020372
|
SLC22A17
|
solute carrier family 22, member 17
|
|
chr10_-_52383611
|
1.134
|
NM_147156
|
SGMS1
|
sphingomyelin synthase 1
|
|
chr7_+_65338349
|
1.126
|
|
VKORC1L1
|
vitamin K epoxide reductase complex, subunit 1-like 1
|
|
chr3_-_133969563
|
1.125
|
NM_001005861
NM_002958
|
RYK
|
RYK receptor-like tyrosine kinase
|
|
chr6_+_16129321
|
1.116
|
|
MYLIP
|
myosin regulatory light chain interacting protein
|
|
chr14_+_53019865
|
1.108
|
NM_001099652
|
GPR137C
|
G protein-coupled receptor 137C
|
|
chr9_+_5629110
|
1.107
|
NM_001135920
NM_001206557
NM_020829
|
KIAA1432
|
KIAA1432
|
|
chr1_+_185703470
|
1.100
|
NM_031935
|
HMCN1
|
hemicentin 1
|
|
chr6_+_161412826
|
1.100
|
|
MAP3K4
|
mitogen-activated protein kinase kinase kinase 4
|
|
chr2_+_189156730
|
1.089
|
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1
|
|
chr18_+_72998258
|
1.085
|
|
TSHZ1
|
teashirt zinc finger homeobox 1
|
|
chr16_-_88772732
|
1.080
|
NM_001171815
|
RNF166
|
ring finger protein 166
|
|
chr2_+_231729623
|
1.079
|
|
ITM2C
|
integral membrane protein 2C
|
|
chr2_+_182756665
|
1.072
|
|
SSFA2
|
sperm specific antigen 2
|
|
chr2_+_178257488
|
1.062
|
|
AGPS
|
alkylglycerone phosphate synthase
|
|
chr9_-_84303595
|
1.059
|
NM_005077
|
TLE1
|
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila)
|
|
chr6_-_84419018
|
1.054
|
|
SNAP91
|
synaptosomal-associated protein, 91kDa homolog (mouse)
|
|
chr8_+_74207093
|
1.051
|
|
RDH10
|
retinol dehydrogenase 10 (all-trans)
|
|
chr12_-_76425375
|
1.047
|
NM_007350
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr2_+_231729501
|
1.046
|
NM_001012514
NM_001012516
NM_030926
|
ITM2C
|
integral membrane protein 2C
|
|
chrX_+_118109314
|
1.045
|
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chr10_-_21805715
|
1.026
|
|
C10orf140
|
chromosome 10 open reading frame 140
|
|
chr11_+_59522905
|
1.023
|
|
STX3
|
syntaxin 3
|
|
chrX_-_119694492
|
1.017
|
|
CUL4B
|
cullin 4B
|
|
chr7_+_116312412
|
1.012
|
NM_000245
NM_001127500
|
MET
|
met proto-oncogene (hepatocyte growth factor receptor)
|
|
chr16_-_88772807
|
1.012
|
NM_178841
|
RNF166
|
ring finger protein 166
|
|
chr12_-_31881949
|
1.010
|
|
AMN1
|
antagonist of mitotic exit network 1 homolog (S. cerevisiae)
|
|
chr19_-_31840118
|
0.989
|
NM_020856
|
TSHZ3
|
teashirt zinc finger homeobox 3
|
|
chr2_+_231729661
|
0.984
|
|
ITM2C
|
integral membrane protein 2C
|
|
chr5_+_95067511
|
0.982
|
|
|
|
|
chr11_-_10315680
|
0.975
|
NM_030962
|
SBF2
|
SET binding factor 2
|
|
chr12_+_124069080
|
0.973
|
|
TMED2
|
transmembrane emp24 domain trafficking protein 2
|
|
chr13_-_113242467
|
0.972
|
NM_006322
|
TUBGCP3
|
tubulin, gamma complex associated protein 3
|
|
chr9_+_108006990
|
0.971
|
|
SLC44A1
|
solute carrier family 44, member 1
|
|
chr3_-_187463474
|
0.969
|
NM_001706
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr16_+_68119211
|
0.963
|
NM_004555
NM_173163
NM_173165
|
NFATC3
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3
|
|
chr6_+_147524866
|
0.963
|
|
STXBP5
|
syntaxin binding protein 5 (tomosyn)
|
|
chr6_+_43138919
|
0.961
|
NM_003131
|
SRF
|
serum response factor (c-fos serum response element-binding transcription factor)
|
|
chr6_-_84418705
|
0.956
|
NM_001242792
|
SNAP91
|
synaptosomal-associated protein, 91kDa homolog (mouse)
|
|
chr9_-_85677784
|
0.954
|
|
RASEF
|
RAS and EF-hand domain containing
|
|
chr1_+_155147051
|
0.951
|
|
TRIM46
|
tripartite motif containing 46
|
|
chr16_-_23521719
|
0.946
|
|
GGA2
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 2
|
|
chr8_+_37654395
|
0.944
|
NM_032777
|
GPR124
|
G protein-coupled receptor 124
|
|
chr19_-_16582810
|
0.944
|
|
EPS15L1
|
epidermal growth factor receptor pathway substrate 15-like 1
|
|
chr2_+_203241044
|
0.941
|
NM_001204
|
BMPR2
|
bone morphogenetic protein receptor, type II (serine/threonine kinase)
|
|
chr1_+_211432732
|
0.933
|
|
RCOR3
|
REST corepressor 3
|
|
chr14_-_23822025
|
0.933
|
NM_016609
|
SLC22A17
|
solute carrier family 22, member 17
|
|
chr12_-_31882006
|
0.930
|
NM_001113402
|
AMN1
|
antagonist of mitotic exit network 1 homolog (S. cerevisiae)
|
|
chr6_-_75994496
|
0.927
|
|
TMEM30A
|
transmembrane protein 30A
|
|
chr2_+_113033146
|
0.923
|
NM_198581
|
ZC3H6
|
zinc finger CCCH-type containing 6
|
|
chr2_+_178257371
|
0.922
|
NM_003659
|
AGPS
|
alkylglycerone phosphate synthase
|
|
chr13_+_58205943
|
0.921
|
|
PCDH17
|
protocadherin 17
|
|
chr5_+_42423811
|
0.917
|
NM_000163
|
GHR
|
growth hormone receptor
|
|
chr13_-_37494370
|
0.916
|
NM_001127217
NM_005905
|
SMAD9
|
SMAD family member 9
|
|
chr6_+_16143281
|
0.915
|
|
MYLIP
|
myosin regulatory light chain interacting protein
|
|
chr5_-_148931001
|
0.909
|
NM_001025105
NM_001892
|
CSNK1A1
|
casein kinase 1, alpha 1
|
|
chr2_+_238536183
|
0.904
|
NM_001137550
|
LRRFIP1
|
leucine rich repeat (in FLII) interacting protein 1
|
|
chr16_+_53164784
|
0.900
|
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr4_-_174450529
|
0.896
|
|
HAND2
|
heart and neural crest derivatives expressed 2
|
|
chr17_+_39382878
|
0.890
|
NM_031961
|
KRTAP9-2
|
keratin associated protein 9-2
|
|
chr17_+_25621105
|
0.890
|
NM_015626
NM_134265
|
WSB1
|
WD repeat and SOCS box containing 1
|
|
chr3_+_32280154
|
0.879
|
NM_178868
|
CMTM8
|
CKLF-like MARVEL transmembrane domain containing 8
|
|
chr5_+_139060186
|
0.875
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr8_-_57123827
|
0.871
|
|
PLAG1
|
pleiomorphic adenoma gene 1
|
|
chr6_+_2766228
|
0.869
|
|
WRNIP1
|
Werner helicase interacting protein 1
|
|
chr6_-_75994477
|
0.864
|
|
TMEM30A
|
transmembrane protein 30A
|
|
chr13_+_49549953
|
0.863
|
NM_001079673
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr9_+_102584136
|
0.863
|
NM_006981
NM_173199
NM_173200
|
NR4A3
|
nuclear receptor subfamily 4, group A, member 3
|
|
chr3_-_88108047
|
0.861
|
NM_001008390
NM_003663
|
CGGBP1
|
CGG triplet repeat binding protein 1
|
|
chr17_+_59477473
|
0.860
|
|
TBX2
|
T-box 2
|
|
chr13_-_77460432
|
0.857
|
NM_138444
|
KCTD12
|
potassium channel tetramerisation domain containing 12
|
|
chr5_+_176560056
|
0.855
|
NM_172349
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr14_+_65879439
|
0.855
|
|
FUT8
|
fucosyltransferase 8 (alpha (1,6) fucosyltransferase)
|
|
chr3_+_32280211
|
0.851
|
|
CMTM8
|
CKLF-like MARVEL transmembrane domain containing 8
|
|
chr8_-_93115453
|
0.849
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chrX_-_10535476
|
0.844
|
NM_001193278
NM_001193279
NM_001193280
NM_001193281
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr1_+_211432726
|
0.842
|
|
RCOR3
|
REST corepressor 3
|
|
chr5_+_92920592
|
0.840
|
|
NR2F1
NR2F2
|
nuclear receptor subfamily 2, group F, member 1
nuclear receptor subfamily 2, group F, member 2
|
|
chr15_-_69113213
|
0.838
|
|
ANP32A
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A
|
|
chr8_-_95274520
|
0.830
|
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr1_+_2005083
|
0.829
|
NM_001033581
|
PRKCZ
|
protein kinase C, zeta
|
|
chr1_+_66999805
|
0.829
|
NM_032291
|
SGIP1
|
SH3-domain GRB2-like (endophilin) interacting protein 1
|
|
chr1_+_84767288
|
0.826
|
NM_001134663
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr17_+_65821625
|
0.825
|
NM_004459
NM_182641
|
BPTF
|
bromodomain PHD finger transcription factor
|
|
chr14_-_77494974
|
0.825
|
NM_024496
|
IRF2BPL
|
interferon regulatory factor 2 binding protein-like
|
|
chr10_-_64576123
|
0.824
|
NM_000399
NM_001136177
NM_001136179
|
EGR2
|
early growth response 2
|
|
chr8_-_95274540
|
0.822
|
NM_005261
NM_181702
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr2_-_193059249
|
0.822
|
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chrX_+_77166178
|
0.821
|
NM_000052
|
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide
|
|
chr5_-_107006585
|
0.821
|
NM_001962
|
EFNA5
|
ephrin-A5
|
|
chr1_+_32757702
|
0.820
|
|
HDAC1
|
histone deacetylase 1
|
|
chr12_+_32260084
|
0.817
|
NM_001003398
NM_001714
|
BICD1
|
bicaudal D homolog 1 (Drosophila)
|
|
chr6_+_1611544
|
0.812
|
|
FOXC1
|
forkhead box C1
|
|
chr2_+_170683900
|
0.812
|
NM_172070
|
UBR3
|
ubiquitin protein ligase E3 component n-recognin 3 (putative)
|
|
chr17_+_25621141
|
0.811
|
|
WSB1
|
WD repeat and SOCS box containing 1
|
|
chr10_+_76586299
|
0.809
|
NM_012330
|
KAT6B
|
K(lysine) acetyltransferase 6B
|
|
chr10_-_64576074
|
0.802
|
|
EGR2
|
early growth response 2
|
|
chr4_-_146859960
|
0.800
|
|
|
|
|
chr11_-_10830034
|
0.798
|
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2
|
|
chr4_+_108852671
|
0.795
|
NM_183075
|
CYP2U1
|
cytochrome P450, family 2, subfamily U, polypeptide 1
|