|
chr18_+_77155927
|
7.710
|
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr18_+_77155714
|
7.664
|
NM_006162
NM_172388
NM_172390
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr22_+_51113069
|
7.445
|
NM_001080420
|
SHANK3
|
SH3 and multiple ankyrin repeat domains 3
|
|
chr18_+_77155966
|
7.095
|
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr3_+_20081853
|
6.545
|
|
KAT2B
|
K(lysine) acetyltransferase 2B
|
|
chr18_+_77155943
|
6.265
|
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr4_-_16900256
|
6.203
|
|
LDB2
|
LIM domain binding 2
|
|
chr3_+_20081523
|
6.167
|
NM_003884
|
KAT2B
|
K(lysine) acetyltransferase 2B
|
|
chr4_-_16900348
|
6.066
|
NM_001130834
NM_001290
|
LDB2
|
LIM domain binding 2
|
|
chr1_+_164528586
|
5.946
|
NM_001204961
NM_001204963
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr4_-_16900028
|
5.745
|
|
LDB2
|
LIM domain binding 2
|
|
chr12_+_123319977
|
5.511
|
NM_003959
|
HIP1R
|
huntingtin interacting protein 1 related
|
|
chr2_+_48757292
|
5.342
|
NM_001198595
NM_006873
|
STON1
|
stonin 1
|
|
chr20_+_42543417
|
4.966
|
NM_001098797
|
TOX2
|
TOX high mobility group box family member 2
|
|
chr9_+_139606916
|
4.938
|
NM_152421
|
FAM69B
|
family with sequence similarity 69, member B
|
|
chr19_-_291335
|
4.906
|
NM_003712
NM_177526
|
PPAP2C
|
phosphatidic acid phosphatase type 2C
|
|
chr14_-_91526912
|
4.817
|
NM_004755
NM_182398
|
RPS6KA5
|
ribosomal protein S6 kinase, 90kDa, polypeptide 5
|
|
chr4_+_100871625
|
4.740
|
|
LOC256880
|
uncharacterized LOC256880
|
|
chr19_-_291168
|
4.659
|
NM_177543
|
PPAP2C
|
phosphatidic acid phosphatase type 2C
|
|
chr17_+_81037504
|
4.603
|
NM_001004431
|
METRNL
|
meteorin, glial cell differentiation regulator-like
|
|
chr17_-_47841517
|
4.565
|
NM_030802
|
FAM117A
|
family with sequence similarity 117, member A
|
|
chr3_+_110790460
|
4.456
|
NM_001243286
NM_015480
|
PVRL3
|
poliovirus receptor-related 3
|
|
chr1_+_210406194
|
4.308
|
NM_019605
|
SERTAD4
|
SERTA domain containing 4
|
|
chr5_+_139028483
|
4.062
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr12_+_3068442
|
3.946
|
NM_003213
NM_201441
NM_201443
|
TEAD4
|
TEA domain family member 4
|
|
chr10_+_8096633
|
3.690
|
NM_001002295
NM_002051
|
GATA3
|
GATA binding protein 3
|
|
chr10_+_8096772
|
3.626
|
|
GATA3
|
GATA binding protein 3
|
|
chr6_+_15245702
|
3.611
|
|
JARID2
|
jumonji, AT rich interactive domain 2
|
|
chr19_-_3028901
|
3.585
|
NM_003260
|
TLE2
|
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila)
|
|
chr13_-_72441312
|
3.577
|
NM_004392
NM_080759
NM_080760
|
DACH1
|
dachshund homolog 1 (Drosophila)
|
|
chr5_+_139028025
|
3.406
|
NM_016463
|
CXXC5
|
CXXC finger protein 5
|
|
chr19_+_35760002
|
3.354
|
|
USF2
|
upstream transcription factor 2, c-fos interacting
|
|
chr3_+_58223228
|
3.340
|
NM_020676
|
ABHD6
|
abhydrolase domain containing 6
|
|
chr3_+_37903643
|
3.334
|
NM_001008392
NM_005808
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like
|
|
chrX_+_66763868
|
3.331
|
NM_000044
|
AR
|
androgen receptor
|
|
chrX_-_119694871
|
3.329
|
|
CUL4B
|
cullin 4B
|
|
chr1_+_61547625
|
3.283
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr17_-_79008372
|
3.269
|
|
FLJ90757
|
uncharacterized LOC440465
|
|
chr3_+_58223415
|
3.268
|
|
ABHD6
|
abhydrolase domain containing 6
|
|
chr17_-_47841468
|
3.226
|
|
FAM117A
|
family with sequence similarity 117, member A
|
|
chr10_+_112631675
|
3.206
|
|
PDCD4
|
programmed cell death 4 (neoplastic transformation inhibitor)
|
|
chr4_+_6271588
|
3.182
|
|
WFS1
|
Wolfram syndrome 1 (wolframin)
|
|
chr1_+_87797371
|
3.181
|
|
LMO4
|
LIM domain only 4
|
|
chr2_-_43453146
|
3.170
|
|
ZFP36L2
|
zinc finger protein 36, C3H type-like 2
|
|
chr7_-_143599163
|
3.162
|
NM_014719
NM_001206938
|
FAM115A
|
family with sequence similarity 115, member A
|
|
chr15_-_65067696
|
3.142
|
NM_194272
|
RBPMS2
|
RNA binding protein with multiple splicing 2
|
|
chr11_-_119234628
|
3.131
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr2_+_24299751
|
3.130
|
|
LOC375190
|
UPF0638 protein B
|
|
chrX_+_9754465
|
3.124
|
NM_001649
|
SHROOM2
|
shroom family member 2
|
|
chr12_-_106532447
|
3.095
|
|
NUAK1
|
NUAK family, SNF1-like kinase, 1
|
|
chr4_+_41362750
|
3.087
|
NM_001112717
NM_001112718
NM_014988
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr14_-_21994342
|
3.074
|
|
SALL2
|
sal-like 2 (Drosophila)
|
|
chr1_+_212458867
|
3.068
|
NM_006243
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha
|
|
chrX_-_70473941
|
3.050
|
|
ZMYM3
|
zinc finger, MYM-type 3
|
|
chr3_-_15900928
|
3.021
|
NM_015199
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr3_+_12838170
|
2.979
|
NM_001162499
NM_012298
|
CAND2
|
cullin-associated and neddylation-dissociated 2 (putative)
|
|
chr7_+_30951414
|
2.952
|
NM_198098
|
AQP1
|
aquaporin 1 (Colton blood group)
|
|
chr12_+_57943806
|
2.912
|
|
KIF5A
|
kinesin family member 5A
|
|
chr1_-_53018630
|
2.900
|
|
ZCCHC11
|
zinc finger, CCHC domain containing 11
|
|
chr6_-_16761600
|
2.896
|
NM_000332
NM_001128164
|
ATXN1
|
ataxin 1
|
|
chr20_-_62680880
|
2.852
|
NM_018419
|
SOX18
|
SRY (sex determining region Y)-box 18
|
|
chr18_+_77160243
|
2.823
|
NM_172387
NM_172389
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr3_-_47620186
|
2.813
|
NM_001206943
NM_001206944
NM_006574
|
CSPG5
|
chondroitin sulfate proteoglycan 5 (neuroglycan C)
|
|
chr22_+_50585483
|
2.812
|
NM_001164106
|
MOV10L1
|
Mov10l1, Moloney leukemia virus 10-like 1, homolog (mouse)
|
|
chr15_-_82338348
|
2.780
|
NM_032246
|
MEX3B
|
mex-3 homolog B (C. elegans)
|
|
chr3_-_18466759
|
2.774
|
NM_001195470
NM_002971
|
SATB1
|
SATB homeobox 1
|
|
chr6_-_29527701
|
2.768
|
NM_006398
|
UBD
|
ubiquitin D
|
|
chr2_+_112656180
|
2.723
|
NM_006343
|
MERTK
|
c-mer proto-oncogene tyrosine kinase
|
|
chrX_-_54522318
|
2.723
|
NM_004463
|
FGD1
|
FYVE, RhoGEF and PH domain containing 1
|
|
chr15_-_82338459
|
2.719
|
|
MEX3B
|
mex-3 homolog B (C. elegans)
|
|
chrX_-_70473979
|
2.713
|
NM_201599
|
ZMYM3
|
zinc finger, MYM-type 3
|
|
chrX_-_119694719
|
2.696
|
NM_001079872
|
CUL4B
|
cullin 4B
|
|
chr3_+_32280154
|
2.689
|
NM_178868
|
CMTM8
|
CKLF-like MARVEL transmembrane domain containing 8
|
|
chr18_+_11981421
|
2.672
|
NM_014214
|
IMPA2
|
inositol(myo)-1(or 4)-monophosphatase 2
|
|
chr9_+_139560193
|
2.669
|
NM_201446
|
EGFL7
|
EGF-like-domain, multiple 7
|
|
chr9_+_139972195
|
2.667
|
|
UAP1L1
|
UDP-N-acteylglucosamine pyrophosphorylase 1-like 1
|
|
chr6_-_79787784
|
2.637
|
|
PHIP
|
pleckstrin homology domain interacting protein
|
|
chr6_-_29527477
|
2.629
|
|
UBD
|
ubiquitin D
|
|
chr11_-_68518910
|
2.629
|
|
MTL5
|
metallothionein-like 5, testis-specific (tesmin)
|
|
chr5_-_171881355
|
2.617
|
|
SH3PXD2B
|
SH3 and PX domains 2B
|
|
chr4_+_6271605
|
2.616
|
|
WFS1
|
Wolfram syndrome 1 (wolframin)
|
|
chr7_-_156685621
|
2.596
|
|
LMBR1
|
limb region 1 homolog (mouse)
|
|
chr7_+_30960914
|
2.595
|
NM_001185060
NM_001185061
NM_001185062
|
AQP1
|
aquaporin 1 (Colton blood group)
|
|
chr9_+_115513050
|
2.592
|
NM_001012994
|
SNX30
|
sorting nexin family member 30
|
|
chr22_-_28197469
|
2.591
|
NM_002430
|
MN1
|
meningioma (disrupted in balanced translocation) 1
|
|
chr9_+_139560243
|
2.580
|
|
EGFL7
|
EGF-like-domain, multiple 7
|
|
chr12_+_9067130
|
2.576
|
NM_004426
|
PHC1
|
polyhomeotic homolog 1 (Drosophila)
|
|
chr7_+_94023872
|
2.567
|
NM_000089
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr2_-_99347430
|
2.530
|
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chr9_+_133971862
|
2.516
|
NM_001185095
NM_001185096
NM_031426
|
AIF1L
|
allograft inflammatory factor 1-like
|
|
chr11_-_130184459
|
2.512
|
NM_014155
|
ZBTB44
|
zinc finger and BTB domain containing 44
|
|
chr3_+_170075449
|
2.506
|
NM_001145098
NM_005414
|
SKIL
|
SKI-like oncogene
|
|
chr5_-_111093251
|
2.482
|
NM_001142479
NM_001142480
NM_001142478
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr4_+_6271558
|
2.476
|
NM_001145853
NM_006005
|
WFS1
|
Wolfram syndrome 1 (wolframin)
|
|
chr14_-_89883306
|
2.457
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr12_+_132379262
|
2.441
|
NM_003565
|
ULK1
|
unc-51-like kinase 1 (C. elegans)
|
|
chr1_+_87794576
|
2.437
|
|
LMO4
|
LIM domain only 4
|
|
chr19_-_31840118
|
2.429
|
NM_020856
|
TSHZ3
|
teashirt zinc finger homeobox 3
|
|
chr13_-_110438896
|
2.420
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr6_+_117803779
|
2.415
|
NM_173674
|
DCBLD1
|
discoidin, CUB and LCCL domain containing 1
|
|
chr18_-_72920989
|
2.391
|
|
|
|
|
chr12_+_3186520
|
2.373
|
NM_001168320
NM_006675
|
TSPAN9
|
tetraspanin 9
|
|
chr4_-_149363498
|
2.335
|
NM_000901
NM_001166104
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2
|
|
chr10_+_76586299
|
2.330
|
NM_012330
|
KAT6B
|
K(lysine) acetyltransferase 6B
|
|
chr3_+_8543508
|
2.330
|
NM_014583
|
LMCD1
|
LIM and cysteine-rich domains 1
|
|
chr2_-_165477818
|
2.318
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr14_+_56584854
|
2.313
|
NM_021255
|
PELI2
|
pellino homolog 2 (Drosophila)
|
|
chr2_-_165477986
|
2.303
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr11_+_59522905
|
2.292
|
|
STX3
|
syntaxin 3
|
|
chr1_-_20812727
|
2.289
|
NM_018584
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr7_+_2559396
|
2.285
|
NM_001040167
NM_001040168
|
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr14_+_56585147
|
2.279
|
|
PELI2
|
pellino homolog 2 (Drosophila)
|
|
chr9_+_130374466
|
2.278
|
NM_001032221
NM_003165
|
STXBP1
|
syntaxin binding protein 1
|
|
chr2_+_235860616
|
2.276
|
NM_014521
|
SH3BP4
|
SH3-domain binding protein 4
|
|
chr10_-_75634219
|
2.268
|
NM_001204492
NM_001222
NM_172169
NM_172170
NM_172171
NM_172173
|
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma
|
|
chr10_-_35103894
|
2.254
|
NM_001184785
NM_001184786
NM_001184787
NM_001184788
NM_001184789
NM_001184790
NM_001184791
NM_001184792
NM_001184793
NM_001184794
NM_019619
|
PARD3
|
par-3 partitioning defective 3 homolog (C. elegans)
|
|
chr19_+_1249882
|
2.246
|
|
MIDN
|
midnolin
|
|
chr1_+_110753313
|
2.246
|
NM_001039574
NM_004978
|
KCNC4
|
potassium voltage-gated channel, Shaw-related subfamily, member 4
|
|
chrX_-_119694492
|
2.243
|
|
CUL4B
|
cullin 4B
|
|
chr9_+_139971938
|
2.236
|
NM_207309
|
UAP1L1
|
UDP-N-acteylglucosamine pyrophosphorylase 1-like 1
|
|
chr12_-_118541655
|
2.233
|
NM_019086
|
VSIG10
|
V-set and immunoglobulin domain containing 10
|
|
chr4_+_1005609
|
2.206
|
NM_001004356
NM_001004358
|
FGFRL1
|
fibroblast growth factor receptor-like 1
|
|
chr14_-_30396811
|
2.197
|
NM_002742
|
PRKD1
|
protein kinase D1
|
|
chr2_+_203499819
|
2.197
|
NM_173511
|
FAM117B
|
family with sequence similarity 117, member B
|
|
chr4_+_1005395
|
2.184
|
|
FGFRL1
|
fibroblast growth factor receptor-like 1
|
|
chr10_-_81205091
|
2.180
|
NM_153367
|
ZCCHC24
|
zinc finger, CCHC domain containing 24
|
|
chr6_-_84419018
|
2.179
|
|
SNAP91
|
synaptosomal-associated protein, 91kDa homolog (mouse)
|
|
chr11_-_33891361
|
2.173
|
NM_001142316
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr2_-_74667553
|
2.168
|
NM_001015056
NM_033046
|
RTKN
|
rhotekin
|
|
chr10_-_126849066
|
2.158
|
NM_001329
|
CTBP2
|
C-terminal binding protein 2
|
|
chr4_-_102268626
|
2.156
|
NM_000944
NM_001130691
NM_001130692
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme
|
|
chr11_-_2906960
|
2.151
|
NM_000076
NM_001122630
NM_001122631
|
CDKN1C
|
cyclin-dependent kinase inhibitor 1C (p57, Kip2)
|
|
chr1_+_65886315
|
2.139
|
NM_001003679
NM_001003680
NM_002303
NM_001198681
|
LEPR
LEPROT
|
leptin receptor
leptin receptor overlapping transcript
|
|
chr14_-_77494621
|
2.131
|
|
IRF2BPL
|
interferon regulatory factor 2 binding protein-like
|
|
chr7_-_27153490
|
2.127
|
|
HOXA3
|
homeobox A3
|
|
chr1_+_15250578
|
2.126
|
NM_001018000
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr2_+_159313466
|
2.123
|
NM_001005476
NM_003628
|
PKP4
|
plakophilin 4
|
|
chr2_-_43453737
|
2.117
|
NM_006887
|
ZFP36L2
|
zinc finger protein 36, C3H type-like 2
|
|
chr9_+_133971902
|
2.106
|
|
AIF1L
|
allograft inflammatory factor 1-like
|
|
chr3_+_50192420
|
2.104
|
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F
|
|
chr9_+_33817213
|
2.095
|
|
UBE2R2
|
ubiquitin-conjugating enzyme E2R 2
|
|
chr11_+_59522801
|
2.091
|
|
STX3
|
syntaxin 3
|
|
chr14_+_96505566
|
2.085
|
NM_001252507
|
C14orf132
|
chromosome 14 open reading frame 132
|
|
chr5_-_81046858
|
2.081
|
|
SSBP2
|
single-stranded DNA binding protein 2
|
|
chr8_+_144816287
|
2.076
|
|
LOC100128338
|
uncharacterized LOC100128338
|
|
chr19_-_19052032
|
2.075
|
NM_004838
|
HOMER3
|
homer homolog 3 (Drosophila)
|
|
chr1_+_93913566
|
2.048
|
NM_001024948
NM_001164473
NM_017737
|
FNBP1L
|
formin binding protein 1-like
|
|
chr1_+_35258598
|
2.047
|
NM_002060
|
GJA4
|
gap junction protein, alpha 4, 37kDa
|
|
chr4_-_102267952
|
2.042
|
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme
|
|
chr5_-_180076572
|
2.042
|
|
FLT4
|
fms-related tyrosine kinase 4
|
|
chr11_-_33891258
|
2.038
|
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr4_-_102268333
|
2.028
|
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme
|
|
chr7_-_156685875
|
2.020
|
NM_022458
|
LMBR1
|
limb region 1 homolog (mouse)
|
|
chr9_-_139581823
|
2.017
|
|
AGPAT2
|
1-acylglycerol-3-phosphate O-acyltransferase 2 (lysophosphatidic acid acyltransferase, beta)
|
|
chr6_-_84418705
|
2.015
|
NM_001242792
|
SNAP91
|
synaptosomal-associated protein, 91kDa homolog (mouse)
|
|
chr12_-_57824787
|
2.013
|
|
R3HDM2
|
R3H domain containing 2
|
|
chr1_+_211432726
|
2.005
|
|
RCOR3
|
REST corepressor 3
|
|
chr3_+_39851028
|
2.004
|
NM_015460
|
MYRIP
|
myosin VIIA and Rab interacting protein
|
|
chr6_-_31865439
|
1.999
|
|
EHMT2
|
euchromatic histone-lysine N-methyltransferase 2
|
|
chr2_-_43453724
|
1.987
|
|
ZFP36L2
|
zinc finger protein 36, C3H type-like 2
|
|
chr10_-_125853102
|
1.985
|
|
CHST15
|
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15
|
|
chr8_+_1711869
|
1.970
|
NM_018941
|
CLN8
|
ceroid-lipofuscinosis, neuronal 8 (epilepsy, progressive with mental retardation)
|
|
chr5_+_139027907
|
1.956
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr18_+_46065367
|
1.935
|
NM_001142397
NM_014772
|
CTIF
|
CBP80/20-dependent translation initiation factor
|
|
chr6_-_29600859
|
1.931
|
|
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1
|
|
chr1_-_3528058
|
1.929
|
NM_001409
|
MEGF6
|
multiple EGF-like-domains 6
|
|
chr5_+_139027955
|
1.929
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr1_+_211432732
|
1.929
|
|
RCOR3
|
REST corepressor 3
|
|
chr4_+_74702272
|
1.926
|
NM_002993
|
CXCL6
|
chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2)
|
|
chr12_+_53443834
|
1.914
|
NM_170754
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr4_-_110650965
|
1.914
|
NM_030821
|
PLA2G12A
|
phospholipase A2, group XIIA
|
|
chr17_-_48943672
|
1.914
|
NM_001243885
NM_005749
|
TOB1
|
transducer of ERBB2, 1
|
|
chr11_-_33890931
|
1.913
|
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr9_-_139581900
|
1.909
|
NM_001012727
NM_006412
|
AGPAT2
|
1-acylglycerol-3-phosphate O-acyltransferase 2 (lysophosphatidic acid acyltransferase, beta)
|
|
chr8_-_22549901
|
1.909
|
NM_001199880
|
EGR3
|
early growth response 3
|
|
chr16_+_29817810
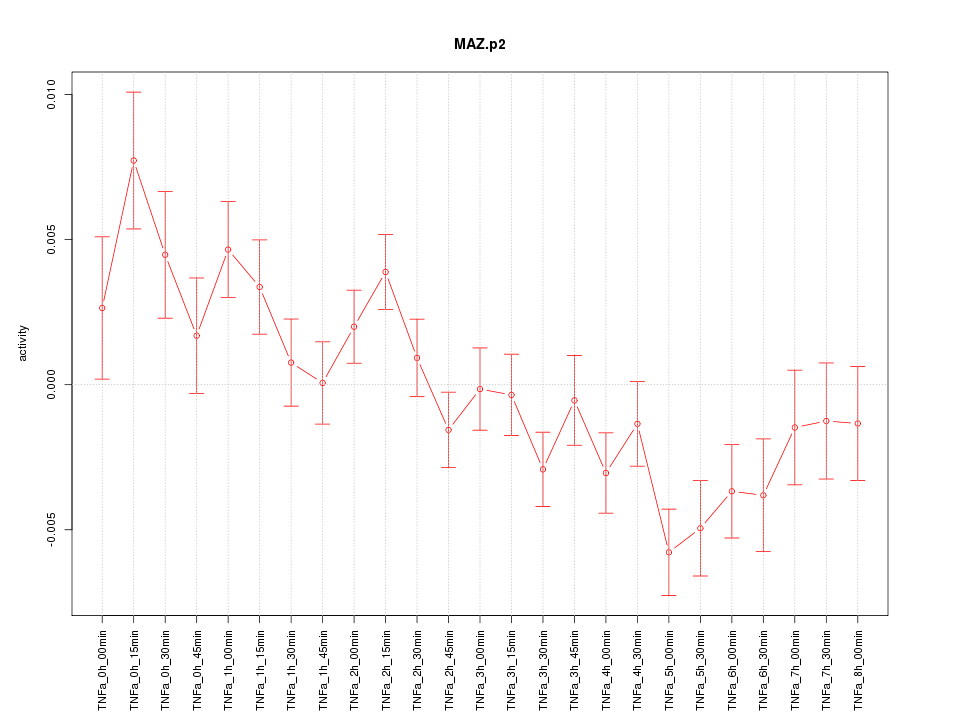
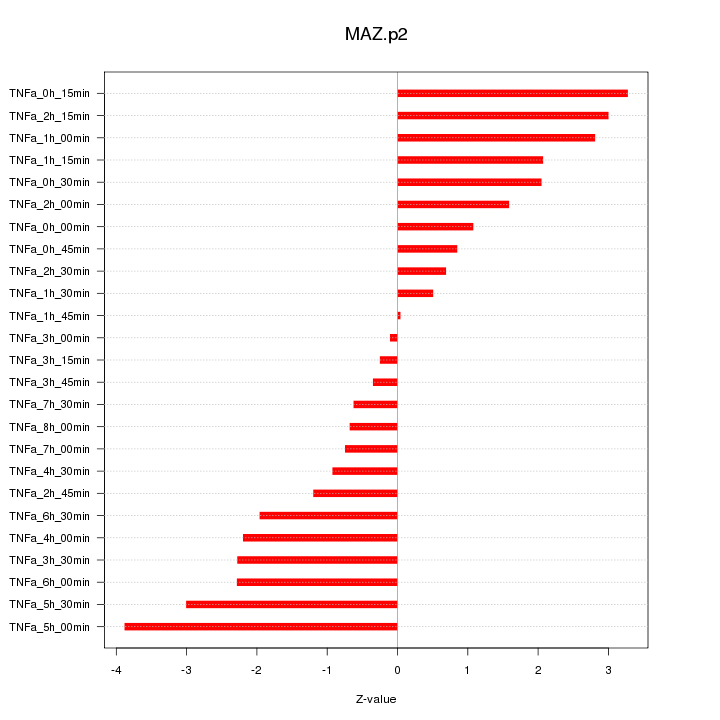
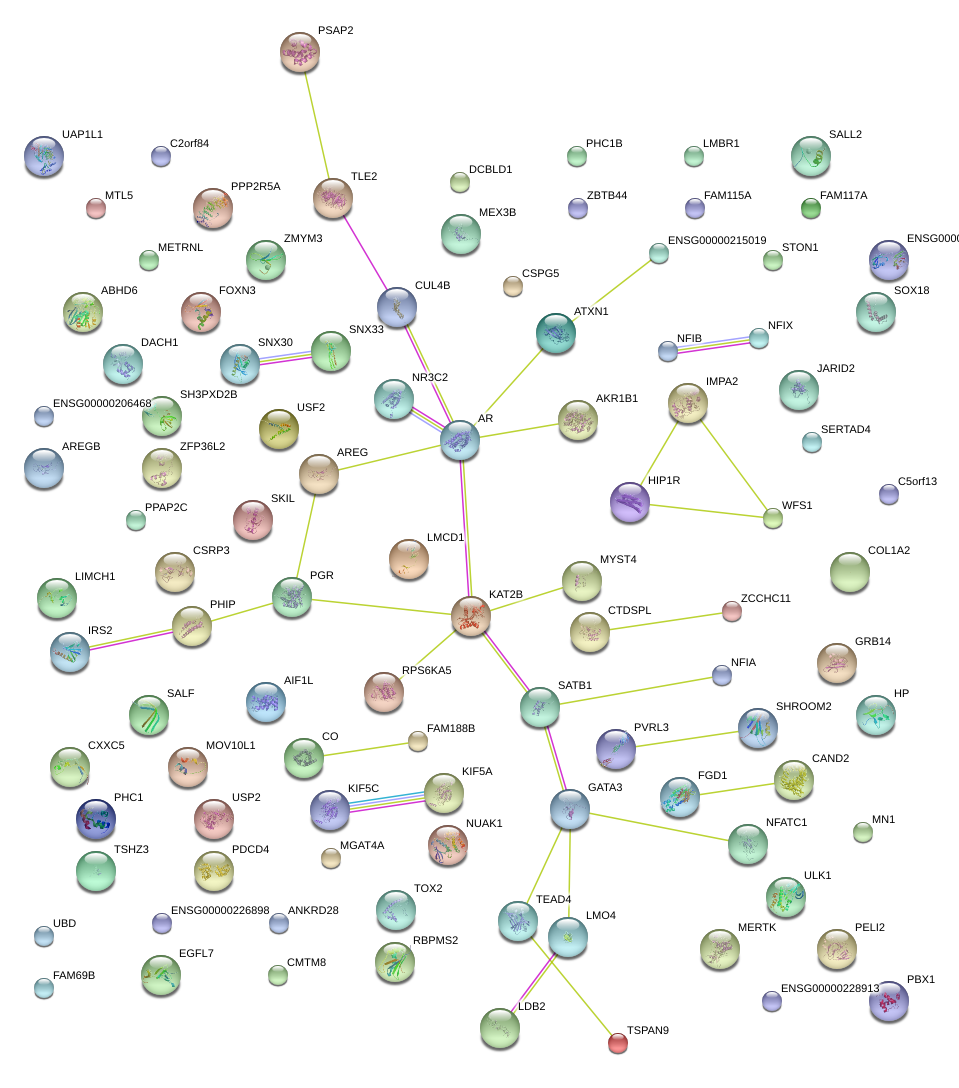
|
1.905
|
NM_001042539
NM_002383
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor)
|
|
chr8_-_74659082
|
1.901
|
NM_001164380
NM_001164381
NM_001164382
NM_001164383
NM_001164385
NM_014393
|
STAU2
|
staufen, RNA binding protein, homolog 2 (Drosophila)
|
|
chr17_+_65821625
|
1.895
|
NM_004459
NM_182641
|
BPTF
|
bromodomain PHD finger transcription factor
|
|
chr8_-_127570465
|
1.889
|
|
FAM84B
|
family with sequence similarity 84, member B
|
|
chr13_-_72440557
|
1.884
|
|
DACH1
|
dachshund homolog 1 (Drosophila)
|
|
chr3_-_169381417
|
1.883
|
NM_001205194
NM_004991
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr5_+_176560882
|
1.878
|
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr6_-_79787887
|
1.871
|
|
PHIP
|
pleckstrin homology domain interacting protein
|
|
chr6_-_144385647
|
1.869
|
NM_001080951
NM_001080955
|
PLAGL1
|
pleiomorphic adenoma gene-like 1
|
|
chr8_-_22550708
|
1.865
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr5_-_114632240
|
1.863
|
NM_001040440
|
CCDC112
|
coiled-coil domain containing 112
|
|
chr1_-_184943673
|
1.856
|
NM_052966
|
FAM129A
|
family with sequence similarity 129, member A
|
|
chr2_+_238600878
|
1.852
|
|
LRRFIP1
|
leucine rich repeat (in FLII) interacting protein 1
|
|
chr10_-_126849556
|
1.849
|
NM_001083914
|
CTBP2
|
C-terminal binding protein 2
|
|
chr4_+_154387450
|
1.847
|
NM_001131007
NM_015196
|
KIAA0922
|
KIAA0922
|
|
chr1_+_222791151
|
1.845
|
NM_198551
|
MIA3
|
melanoma inhibitory activity family, member 3
|
|
chr1_+_211432693
|
1.840
|
NM_001136223
NM_001136224
NM_001136225
|
RCOR3
|
REST corepressor 3
|
|
chr6_-_29600959
|
1.837
|
NM_001470
NM_021904
|
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1
|
|
chr9_+_71320105
|
1.830
|
NM_003558
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta
|
|
chr19_+_55996537
|
1.823
|
NM_020378
|
NAT14
|
N-acetyltransferase 14 (GCN5-related, putative)
|
|
chr8_-_28243589
|
1.821
|
|
ZNF395
|
zinc finger protein 395
|
|
chr8_+_120220554
|
1.819
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr2_+_200323088
|
1.814
|
|
|
|
|
chr7_+_94024201
|
1.809
|
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr9_+_127020242
|
1.803
|
|
NEK6
|
NIMA (never in mitosis gene a)-related kinase 6
|