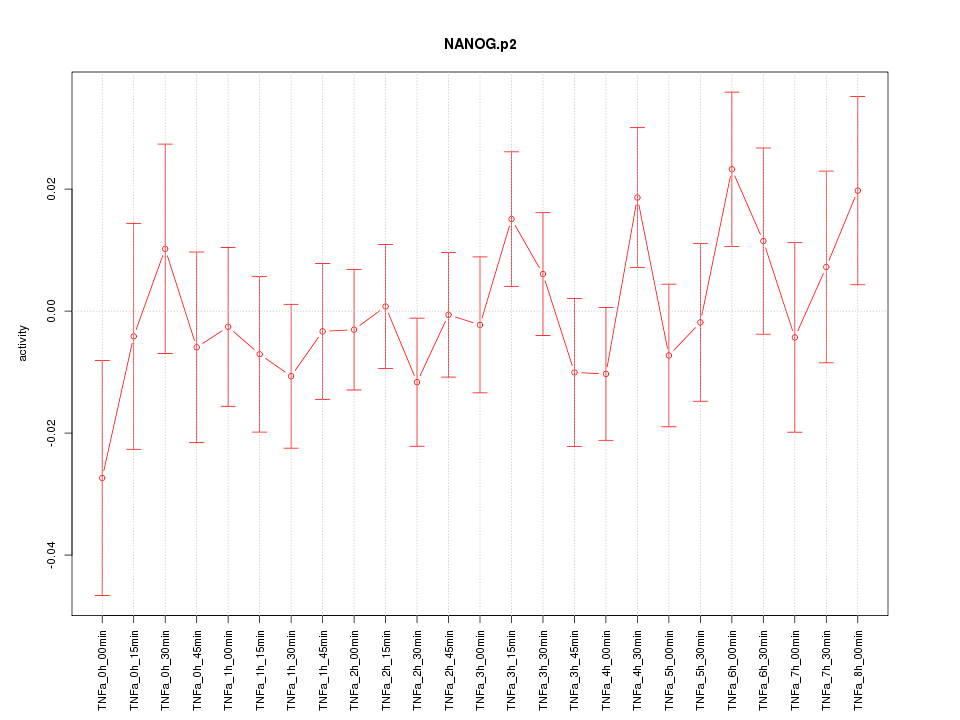
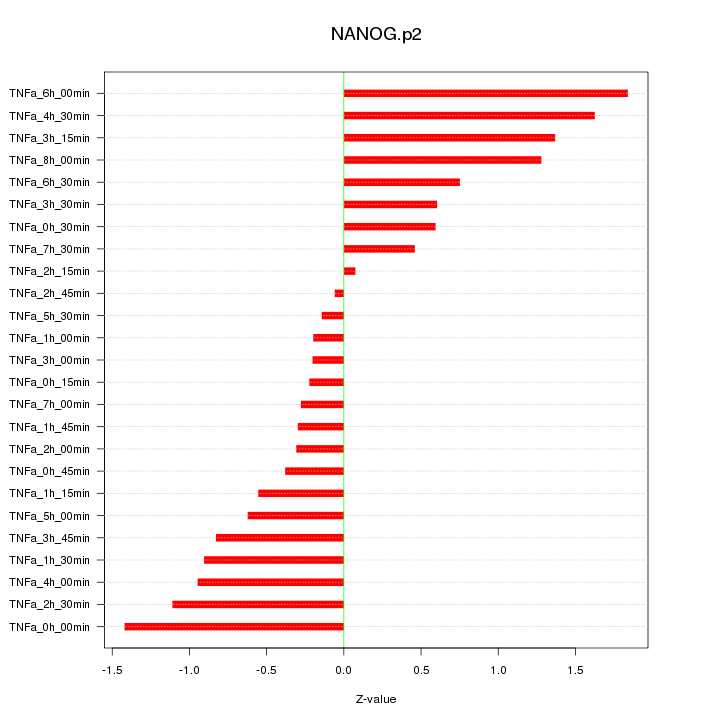
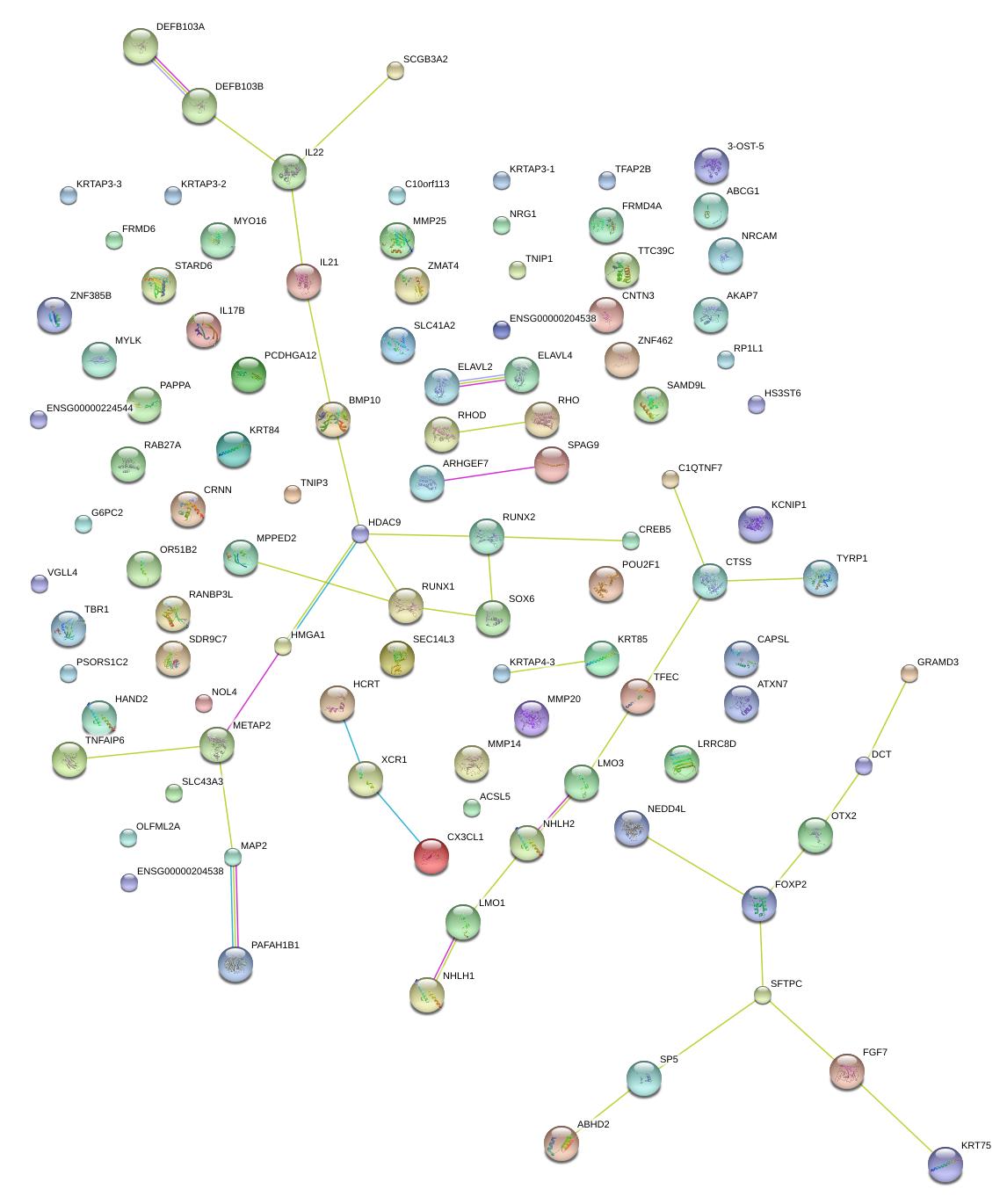
|
chr12_-_52779362
|
3.063
|
NM_033045
|
KRT84
|
keratin 84
|
|
chr5_-_35938736
|
1.565
|
NM_001042625
NM_144647
|
CAPSL
|
calcyphosine-like
|
|
chr12_-_16760935
|
1.391
|
NM_001001395
NM_001243609
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr17_-_39165343
|
1.363
|
NM_031958
|
KRTAP3-1
|
keratin associated protein 3-1
|
|
chr5_-_150467220
|
1.305
|
NM_001252390
|
TNIP1
|
TNFAIP3 interacting protein 1
|
|
chr12_-_105322056
|
1.256
|
NM_032148
|
SLC41A2
|
solute carrier family 41, member 2
|
|
chr2_+_152214105
|
1.246
|
NM_007115
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6
|
|
chr1_+_15990300
|
1.178
|
|
|
|
|
chr11_-_57195052
|
1.051
|
NM_017611
|
SLC43A3
|
solute carrier family 43, member 3
|
|
chr1_-_150738232
|
1.043
|
|
CTSS
|
cathepsin S
|
|
chr18_+_55888750
|
0.986
|
NM_001144966
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr9_+_109625347
|
0.971
|
NM_021224
|
ZNF462
|
zinc finger protein 462
|
|
chr9_+_12693385
|
0.970
|
NM_000550
|
TYRP1
|
tyrosinase-related protein 1
|
|
chr13_+_109281569
|
0.948
|
NM_001198950
|
MYO16
|
myosin XVI
|
|
chr16_+_57406398
|
0.934
|
NM_002996
|
CX3CL1
|
chemokine (C-X3-C motif) ligand 1
|
|
chr3_-_74570262
|
0.917
|
NM_020872
|
CNTN3
|
contactin 3 (plasmacytoma associated)
|
|
chr14_+_51955854
|
0.910
|
NM_001042481
|
FRMD6
|
FERM domain containing 6
|
|
chr18_+_55888799
|
0.894
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr15_-_55562483
|
0.893
|
NM_183234
NM_004580
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr18_-_51880942
|
0.885
|
NM_139171
|
STARD6
|
StAR-related lipid transfer (START) domain containing 6
|
|
chr18_+_55888838
|
0.869
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr9_+_118916069
|
0.849
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr3_-_123339423
|
0.836
|
NM_053031
NM_053032
|
MYLK
|
myosin light chain kinase
|
|
chr10_-_13749622
|
0.830
|
|
FRMD4A
|
FERM domain containing 4A
|
|
chr1_-_150738292
|
0.813
|
NM_001199739
NM_004079
|
CTSS
|
cathepsin S
|
|
chr5_+_140809957
|
0.782
|
|
PCDHGA12
|
protocadherin gamma subfamily A, 12
|
|
chr2_+_162272892
|
0.778
|
|
TBR1
|
T-box, brain, 1
|
|
chr7_-_92777657
|
0.759
|
NM_152703
|
SAMD9L
|
sterile alpha motif domain containing 9-like
|
|
chr17_-_49124128
|
0.754
|
NM_001251971
|
SPAG9
|
sperm associated antigen 9
|
|
chr5_-_148758787
|
0.746
|
NM_014443
|
IL17B
|
interleukin 17B
|
|
chr6_+_34206567
|
0.741
|
NM_145905
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr10_-_14372807
|
0.740
|
NM_018027
|
FRMD4A
|
FERM domain containing 4A
|
|
chr10_+_114133775
|
0.736
|
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr2_-_180427314
|
0.736
|
NM_001113398
|
ZNF385B
|
zinc finger protein 385B
|
|
chr15_-_55563091
|
0.732
|
NM_183236
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr5_+_169780880
|
0.731
|
NM_001034838
|
KCNIP1
|
Kv channel interacting protein 1
|
|
chr3_-_11623829
|
0.727
|
NM_001128220
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr6_+_131571298
|
0.707
|
NM_004842
NM_138633
|
AKAP7
|
A kinase (PRKA) anchor protein 7
|
|
chr1_+_50574601
|
0.705
|
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr2_-_69098502
|
0.704
|
NM_014482
|
BMP10
|
bone morphogenetic protein 10
|
|
chr5_-_36301999
|
0.702
|
NM_001161429
NM_145000
|
RANBP3L
|
RAN binding protein 3-like
|
|
chr12_-_16759939
|
0.698
|
NM_001243613
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr21_+_43710190
|
0.687
|
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1
|
|
chr7_+_28725597
|
0.680
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr17_-_39150384
|
0.676
|
NM_033185
|
KRTAP3-3
|
keratin associated protein 3-3
|
|
chr3_-_123339178
|
0.674
|
|
MYLK
|
myosin light chain kinase
|
|
chr4_-_122137781
|
0.663
|
NM_001128843
|
TNIP3
|
TNFAIP3 interacting protein 3
|
|
chr12_-_16759733
|
0.661
|
NM_001243610
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr11_-_102496049
|
0.660
|
NM_004771
|
MMP20
|
matrix metallopeptidase 20
|
|
chr3_-_46068978
|
0.660
|
NM_001024644
NM_005283
|
XCR1
|
chemokine (C motif) receptor 1
|
|
chr18_+_21572736
|
0.658
|
NM_153211
|
TTC39C
|
tetratricopeptide repeat domain 39C
|
|
chr10_-_21435487
|
0.644
|
NM_001010896
NM_001177483
|
C10orf113
|
chromosome 10 open reading frame 113
|
|
chr14_+_23305816
|
0.640
|
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted)
|
|
chr5_+_125758818
|
0.628
|
NM_001146320
NM_001146321
NM_023927
|
GRAMD3
|
GRAM domain containing 3
|
|
chr7_+_18535351
|
0.617
|
NM_001204145
NM_001204146
NM_014707
NM_178423
|
HDAC9
|
histone deacetylase 9
|
|
chr7_-_107880492
|
0.609
|
NM_001037132
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr18_-_31803168
|
0.608
|
|
NOL4
|
nucleolar protein 4
|
|
chr1_+_167298382
|
0.601
|
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr2_+_169757749
|
0.595
|
NM_001081686
NM_021176
|
G6PC2
|
glucose-6-phosphatase, catalytic, 2
|
|
chr14_-_57272344
|
0.589
|
NM_172337
|
OTX2
|
orthodenticle homeobox 2
|
|
chr21_-_36421586
|
0.579
|
NM_001754
|
RUNX1
|
runt-related transcription factor 1
|
|
chr21_-_36421630
|
0.578
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr17_-_39324423
|
0.577
|
NM_033187
|
KRTAP4-3
|
keratin associated protein 4-3
|
|
chr3_-_123411185
|
0.571
|
|
MYLK
|
myosin light chain kinase
|
|
chr3_+_129247481
|
0.570
|
NM_000539
|
RHO
|
rhodopsin
|
|
chr8_+_32504250
|
0.568
|
NM_013959
|
NRG1
|
neuregulin 1
|
|
chr17_-_40337384
|
0.563
|
NM_001524
|
HCRT
|
hypocretin (orexin) neuropeptide precursor
|
|
chr15_+_49715374
|
0.547
|
NM_002009
|
FGF7
|
fibroblast growth factor 7
|
|
chr6_+_50786438
|
0.542
|
NM_003221
|
TFAP2B
|
transcription factor AP-2 beta (activating enhancer binding protein 2 beta)
|
|
chr18_-_31803514
|
0.532
|
NM_001198546
NM_001198548
NM_003787
|
NOL4
|
nucleolar protein 4
|
|
chr12_-_52828105
|
0.527
|
NM_004693
|
KRT75
|
keratin 75
|
|
chr13_-_95131718
|
0.512
|
NM_001129889
NM_001922
|
DCT
|
dopachrome tautomerase (dopachrome delta-isomerase, tyrosine-related protein 2)
|
|
chr1_-_152386727
|
0.511
|
NM_016190
|
CRNN
|
cornulin
|
|
chr1_+_167298280
|
0.510
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr8_-_7287651
|
0.508
|
NM_018661
|
DEFB103B
|
defensin, beta 103B
|
|
chr7_-_115670774
|
0.507
|
|
TFEC
|
transcription factor EC
|
|
chr3_+_63884074
|
0.502
|
NM_001177387
|
ATXN7
|
ataxin 7
|
|
chr7_+_114055171
|
0.499
|
|
FOXP2
|
forkhead box P2
|
|
chr14_+_23305792
|
0.498
|
NM_004995
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted)
|
|
chr2_+_210444154
|
0.494
|
|
MAP2
|
microtubule-associated protein 2
|
|
chr11_-_16424391
|
0.493
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr10_+_114133838
|
0.489
|
NM_203379
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr5_+_147258273
|
0.488
|
NM_054023
|
SCGB3A2
|
secretoglobin, family 3A, member 2
|
|
chr1_-_116383692
|
0.486
|
NM_005599
|
NHLH2
|
nescient helix loop helix 2
|
|
chr12_-_52761254
|
0.481
|
NM_002283
|
KRT85
|
keratin 85
|
|
chr4_-_123542165
|
0.480
|
NM_001207006
NM_021803
|
IL21
|
interleukin 21
|
|
chr12_-_57328044
|
0.474
|
NM_148897
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C, member 7
|
|
chr7_+_28452143
|
0.471
|
NM_182898
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr4_-_122148620
|
0.468
|
NM_001244764
|
TNIP3
|
TNFAIP3 interacting protein 3
|
|
chr1_+_90286469
|
0.465
|
NM_001134479
|
LRRC8D
|
leucine rich repeat containing 8 family, member D
|
|
chr6_-_31107110
|
0.465
|
NM_014069
|
PSORS1C2
|
psoriasis susceptibility 1 candidate 2
|
|
chr8_+_32579350
|
0.464
|
NM_001159996
|
NRG1
|
neuregulin 1
|
|
chr8_-_40755328
|
0.462
|
NM_001135731
NM_024645
|
ZMAT4
|
zinc finger, matrin-type 4
|
|
chr15_+_89631625
|
0.460
|
|
ABHD2
|
abhydrolase domain containing 2
|
|
chr6_+_34204928
|
0.452
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr8_-_10512616
|
0.448
|
NM_178857
|
RP1L1
|
retinitis pigmentosa 1-like 1
|
|
chr11_-_5345530
|
0.446
|
NM_033180
|
OR51B2
|
olfactory receptor, family 51, subfamily B, member 2
|
|
chr2_+_171571800
|
0.441
|
NM_001003845
|
SP5
|
Sp5 transcription factor
|
|
chr4_+_15429628
|
0.439
|
NM_031911
|
C1QTNF7
|
C1q and tumor necrosis factor related protein 7
|
|
chr4_-_174451356
|
0.435
|
NM_021973
|
HAND2
|
heart and neural crest derivatives expressed 2
|
|
chr17_-_1248346
|
0.434
|
|
PAFAH1B1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa)
|
|
chr22_-_30867944
|
0.430
|
NM_174975
|
SEC14L3
|
SEC14-like 3 (S. cerevisiae)
|
|
chr11_-_30601919
|
0.427
|
NM_001584
|
MPPED2
|
metallophosphoesterase domain containing 2
|
|
chr5_+_140810126
|
0.426
|
NM_003735
NM_032094
|
PCDHGA12
|
protocadherin gamma subfamily A, 12
|
|
chr7_+_28475233
|
0.422
|
NM_004904
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr12_-_16757944
|
0.418
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr6_-_114384008
|
0.409
|
NM_153612
|
HS3ST5
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 5
|
|
chrX_+_50027539
|
0.404
|
NM_033031
NM_033670
|
CCNB3
|
cyclin B3
|
|
chr11_-_57089670
|
0.399
|
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa
|
|
chr7_-_25268057
|
0.398
|
NM_022150
|
NPVF
|
neuropeptide VF precursor
|
|
chr12_-_57505176
|
0.397
|
NM_001178078
NM_001178080
NM_001178081
NM_003153
|
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced
|
|
chr1_-_42384370
|
0.396
|
NM_001127714
NM_024503
|
HIVEP3
|
human immunodeficiency virus type I enhancer binding protein 3
|
|
chr7_+_114066554
|
0.394
|
NM_001172767
NM_148899
|
FOXP2
|
forkhead box P2
|
|
chr1_+_160160284
|
0.391
|
NM_001231
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle)
|
|
chr4_+_169013687
|
0.390
|
NM_007193
|
ANXA10
|
annexin A10
|
|
chr3_+_152880024
|
0.389
|
NM_002886
|
RAP2B
|
RAP2B, member of RAS oncogene family
|
|
chr7_+_114055048
|
0.387
|
NM_001172766
NM_014491
NM_148898
NM_148900
|
FOXP2
|
forkhead box P2
|
|
chr1_-_150738255
|
0.382
|
|
CTSS
|
cathepsin S
|
|
chr13_-_99910548
|
0.382
|
NM_001098200
NM_005292
|
GPR18
|
G protein-coupled receptor 18
|
|
chr6_+_34204690
|
0.378
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr5_+_170736287
|
0.377
|
NM_021025
|
TLX3
|
T-cell leukemia homeobox 3
|
|
chr11_-_117747618
|
0.377
|
NM_001204268
NM_001243598
NM_001164832
NM_001164836
NM_001164837
NM_022003
|
FXYD6-FXYD2
FXYD6
|
FXYD6-FXYD2 readthrough
FXYD domain containing ion transport regulator 6
|
|
chr1_-_169703178
|
0.376
|
NM_000450
|
SELE
|
selectin E
|
|
chr2_-_55199894
|
0.375
|
|
|
|
|
chr5_+_140810181
|
0.374
|
|
PCDHGA12
|
protocadherin gamma subfamily A, 12
|
|
chr1_+_74701070
|
0.374
|
NM_015978
|
TNNI3K
|
TNNI3 interacting kinase
|
|
chr2_-_163008913
|
0.374
|
NM_002054
|
GCG
|
glucagon
|
|
chr11_+_75526211
|
0.362
|
NM_003369
|
UVRAG
|
UV radiation resistance associated gene
|
|
chr1_+_50569581
|
0.362
|
NM_001144776
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr16_+_56815703
|
0.360
|
NM_001242795
|
NUP93
|
nucleoporin 93kDa
|
|
chr14_-_95236351
|
0.359
|
NM_173849
|
GSC
|
goosecoid homeobox
|
|
chr8_+_24298442
|
0.356
|
NM_003817
|
ADAM7
|
ADAM metallopeptidase domain 7
|
|
chr6_-_42690308
|
0.355
|
NM_000322
|
PRPH2
|
peripherin 2 (retinal degeneration, slow)
|
|
chr10_-_52645384
|
0.353
|
|
A1CF
|
APOBEC1 complementation factor
|
|
chr11_-_117748200
|
0.352
|
NM_001164831
|
FXYD6
|
FXYD domain containing ion transport regulator 6
|
|
chr14_-_38064440
|
0.349
|
NM_004496
|
FOXA1
|
forkhead box A1
|
|
chr9_-_86584079
|
0.346
|
|
HNRNPK
|
heterogeneous nuclear ribonucleoprotein K
|
|
chr20_-_30539725
|
0.346
|
|
PDRG1
|
p53 and DNA-damage regulated 1
|
|
chr3_+_35721115
|
0.340
|
NM_001025068
NM_001025069
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr22_-_40859423
|
0.339
|
|
MKL1
|
megakaryoblastic leukemia (translocation) 1
|
|
chr2_-_180610768
|
0.338
|
NM_001113397
|
ZNF385B
|
zinc finger protein 385B
|
|
chr8_+_54764367
|
0.336
|
NM_170587
|
RGS20
|
regulator of G-protein signaling 20
|
|
chr5_-_169816637
|
0.335
|
NM_004137
|
KCNMB1
|
potassium large conductance calcium-activated channel, subfamily M, beta member 1
|
|
chr9_+_130026755
|
0.334
|
NM_032293
|
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3
|
|
chr11_-_128712340
|
0.327
|
NM_000220
|
KCNJ1
|
potassium inwardly-rectifying channel, subfamily J, member 1
|
|
chr7_+_129906702
|
0.326
|
NM_001869
|
CPA2
|
carboxypeptidase A2 (pancreatic)
|
|
chr1_-_200377488
|
0.325
|
|
ZNF281
|
zinc finger protein 281
|
|
chr18_+_32173281
|
0.320
|
NM_001128175
NM_001198938
NM_001198939
NM_001198940
|
DTNA
|
dystrobrevin, alpha
|
|
chr12_-_16759531
|
0.318
|
NM_018640
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr8_+_86157693
|
0.316
|
NM_198584
|
CA13
|
carbonic anhydrase XIII
|
|
chr4_-_159956332
|
0.315
|
NM_152543
|
C4orf45
|
chromosome 4 open reading frame 45
|
|
chr12_-_57505121
|
0.315
|
|
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced
|
|
chr11_-_85430117
|
0.314
|
NM_206929
|
SYTL2
|
synaptotagmin-like 2
|
|
chr7_+_129984629
|
0.313
|
NM_001127441
NM_001127442
NM_080385
|
CPA5
|
carboxypeptidase A5
|
|
chr3_-_101039403
|
0.313
|
NM_016247
|
IMPG2
|
interphotoreceptor matrix proteoglycan 2
|
|
chr1_+_209878135
|
0.313
|
NM_005525
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1
|
|
chr5_+_140810326
|
0.312
|
|
PCDHGA12
|
protocadherin gamma subfamily A, 12
|
|
chr1_-_178840079
|
0.312
|
NM_004673
|
ANGPTL1
|
angiopoietin-like 1
|
|
chr7_-_115670828
|
0.312
|
NM_001018058
NM_012252
|
TFEC
|
transcription factor EC
|
|
chr11_+_75526236
|
0.312
|
|
UVRAG
|
UV radiation resistance associated gene
|
|
chr2_-_209028288
|
0.310
|
NM_014617
|
CRYGA
|
crystallin, gamma A
|
|
chr1_-_200377262
|
0.310
|
|
ZNF281
|
zinc finger protein 281
|
|
chr3_+_115342150
|
0.309
|
NM_001130064
NM_002045
|
GAP43
|
growth associated protein 43
|
|
chr12_-_4488707
|
0.307
|
NM_020638
|
FGF23
|
fibroblast growth factor 23
|
|
chr20_+_43030005
|
0.306
|
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr3_+_88188261
|
0.305
|
NM_018293
|
ZNF654
|
zinc finger protein 654
|
|
chr15_+_66694216
|
0.305
|
|
MAP2K1
|
mitogen-activated protein kinase kinase 1
|
|
chr11_-_58980321
|
0.303
|
NM_001039396
|
MPEG1
|
macrophage expressed 1
|
|
chr2_-_49381555
|
0.301
|
NM_000145
NM_181446
|
FSHR
|
follicle stimulating hormone receptor
|
|
chr9_+_109686436
|
0.299
|
|
ZNF462
|
zinc finger protein 462
|
|
chr2_+_36923902
|
0.299
|
|
VIT
|
vitrin
|
|
chr16_+_6823809
|
0.297
|
NM_001142334
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr3_+_121796695
|
0.294
|
NM_006889
NM_176892
|
CD86
|
CD86 molecule
|
|
chr5_+_125759076
|
0.293
|
|
GRAMD3
|
GRAM domain containing 3
|
|
chr5_+_34687663
|
0.293
|
NM_001145523
|
RAI14
|
retinoic acid induced 14
|
|
chr16_-_73092533
|
0.293
|
NM_001164766
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr15_+_89631731
|
0.292
|
|
ABHD2
|
abhydrolase domain containing 2
|
|
chr12_-_57504935
|
0.292
|
|
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced
|
|
chr21_-_31538970
|
0.291
|
NM_012131
|
CLDN17
|
claudin 17
|
|
chr8_-_20040612
|
0.290
|
NM_001135691
NM_001142324
NM_001142325
NM_003053
|
SLC18A1
|
solute carrier family 18 (vesicular monoamine), member 1
|
|
chr2_+_171784947
|
0.285
|
NM_001201428
NM_015530
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa
|
|
chr17_+_39240412
|
0.283
|
NM_033061
|
KRTAP4-7
|
keratin associated protein 4-7
|
|
chr6_-_46293628
|
0.279
|
NM_005822
|
RCAN2
|
regulator of calcineurin 2
|
|
chr3_+_115342302
|
0.277
|
|
GAP43
|
growth associated protein 43
|
|
chr12_-_53045947
|
0.276
|
NM_000423
|
KRT2
|
keratin 2
|
|
chr1_+_50575465
|
0.276
|
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr22_-_17747520
|
0.276
|
|
CECR3
|
cat eye syndrome chromosome region, candidate 3 (non-protein coding)
|
|
chr11_+_124481451
|
0.275
|
NM_052959
|
PANX3
|
pannexin 3
|
|
chr5_+_155753755
|
0.273
|
NM_000337
NM_001128209
NM_172244
|
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein)
|
|
chr10_+_18549583
|
0.269
|
NM_000724
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr19_-_39235113
|
0.268
|
NM_144691
|
CAPN12
|
calpain 12
|
|
chr18_+_76740274
|
0.267
|
NM_171999
|
SALL3
|
sal-like 3 (Drosophila)
|
|
chr6_-_46293404
|
0.266
|
|
RCAN2
|
regulator of calcineurin 2
|
|
chr7_+_129015483
|
0.265
|
NM_001130722
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr2_-_37374948
|
0.261
|
NM_001135652
|
EIF2AK2
|
eukaryotic translation initiation factor 2-alpha kinase 2
|
|
chr14_+_32798478
|
0.260
|
NM_004274
|
AKAP6
|
A kinase (PRKA) anchor protein 6
|
|
chr8_-_12612968
|
0.260
|
NM_152271
|
LONRF1
|
LON peptidase N-terminal domain and ring finger 1
|
|
chr12_-_16759430
|
0.259
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr6_+_64231626
|
0.257
|
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr11_+_131240370
|
0.257
|
NM_001048209
|
NTM
|
neurotrimin
|