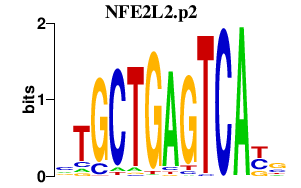
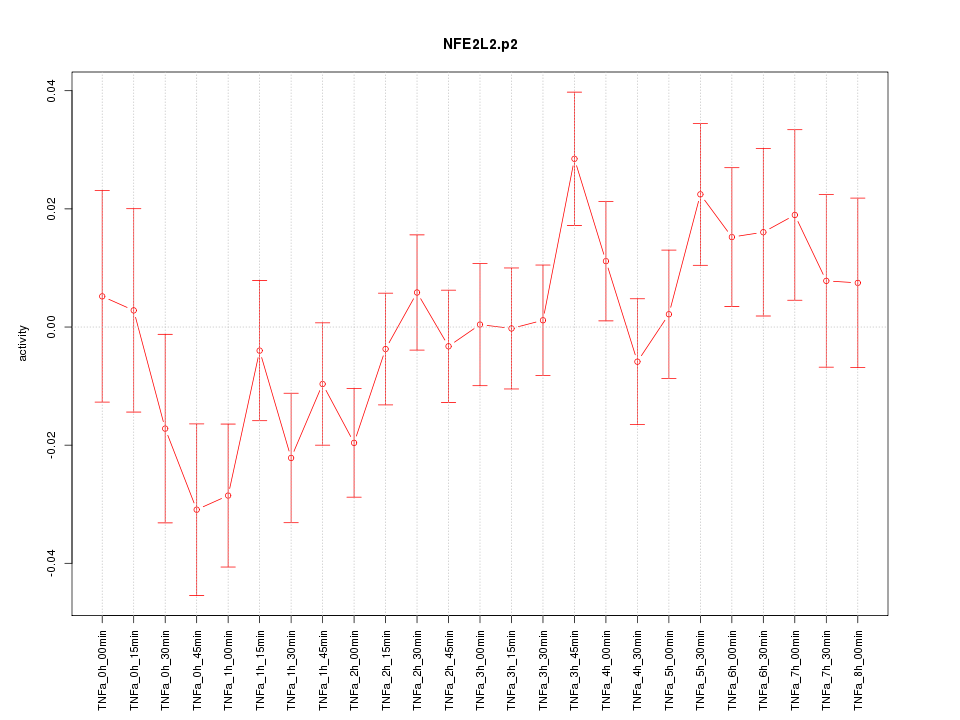
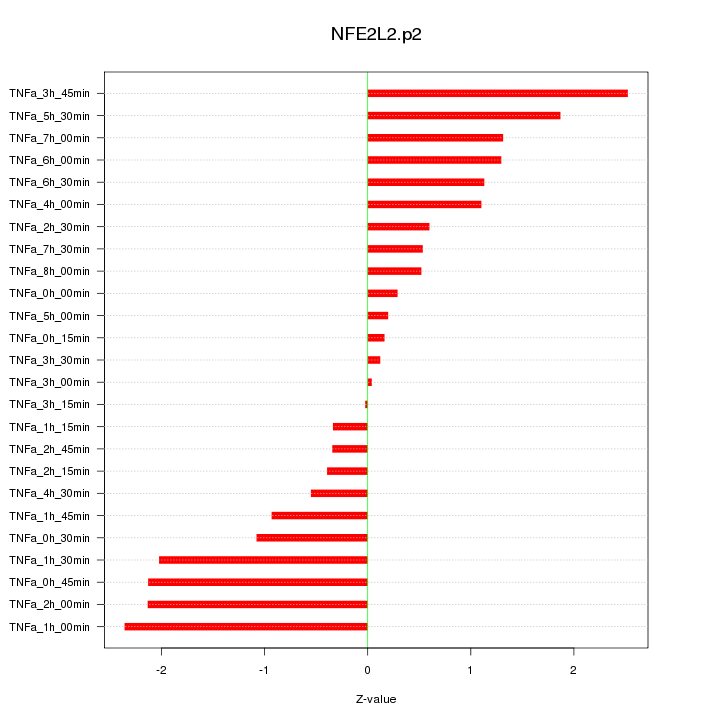
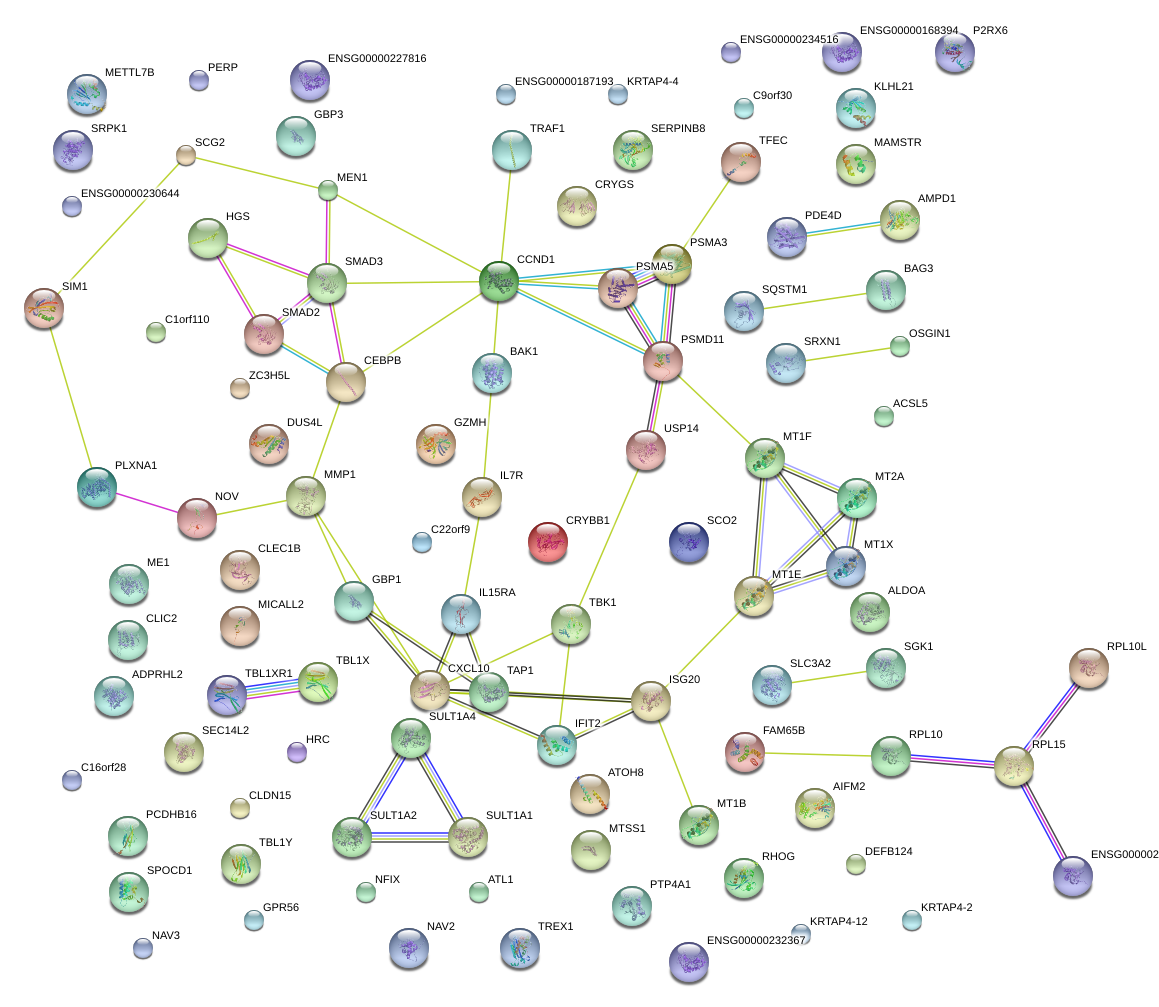
|
chr10_-_6019880
|
5.803
|
NM_001243539
|
IL15RA
|
interleukin 15 receptor, alpha
|
|
chr10_-_6019454
|
5.472
|
NM_002189
NM_172200
|
IL15RA
|
interleukin 15 receptor, alpha
|
|
chr9_-_123688352
|
4.812
|
|
TRAF1
|
TNF receptor-associated factor 1
|
|
chr4_-_76944585
|
4.643
|
NM_001565
|
CXCL10
|
chemokine (C-X-C motif) ligand 10
|
|
chr15_+_89182038
|
3.636
|
NM_002201
|
ISG20
|
interferon stimulated exonuclease gene 20kDa
|
|
chr11_-_102668822
|
3.463
|
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase)
|
|
chr5_+_35856990
|
3.382
|
NM_002185
|
IL7R
|
interleukin 7 receptor
|
|
chr12_+_56075329
|
2.875
|
NM_152637
|
METTL7B
|
methyltransferase like 7B
|
|
chr12_+_56075418
|
2.800
|
|
METTL7B
|
methyltransferase like 7B
|
|
chr6_-_32821743
|
2.379
|
NM_000593
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP)
|
|
chr11_-_102668892
|
2.354
|
NM_001145938
NM_002421
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase)
|
|
chr16_+_57662301
|
2.264
|
NM_001145771
NM_001145772
NM_001145773
NM_201524
NM_201525
|
GPR56
|
G protein-coupled receptor 56
|
|
chr12_-_10151689
|
2.238
|
NM_001099431
NM_016509
|
CLEC1B
|
C-type lectin domain family 1, member B
|
|
chr7_-_115670774
|
2.236
|
|
TFEC
|
transcription factor EC
|
|
chrX_-_154563735
|
2.174
|
NM_001289
|
CLIC2
|
chloride intracellular channel 2
|
|
chr1_-_89488412
|
2.084
|
NM_018284
|
GBP3
|
guanylate binding protein 3
|
|
chr12_+_78225068
|
2.045
|
NM_014903
|
NAV3
|
neuron navigator 3
|
|
chr6_-_138428477
|
2.021
|
NM_022121
|
PERP
|
PERP, TP53 apoptosis effector
|
|
chr1_+_36554452
|
1.994
|
NM_017825
|
ADPRHL2
|
ADP-ribosylhydrolase like 2
|
|
chr6_-_84140854
|
1.700
|
NM_002395
|
ME1
|
malic enzyme 1, NADP(+)-dependent, cytosolic
|
|
chr7_-_1498961
|
1.694
|
|
MICALL2
|
MICAL-like 2
|
|
chrX_+_9431314
|
1.693
|
NM_001139466
NM_001139467
NM_001139468
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr19_-_49222956
|
1.659
|
NM_001130915
|
MAMSTR
|
MEF2 activating motif and SAP domain containing transcriptional regulator
|
|
chr17_+_79650961
|
1.620
|
NM_004712
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate
|
|
chr7_-_115670828
|
1.612
|
NM_001018058
NM_012252
|
TFEC
|
transcription factor EC
|
|
chr6_-_84140744
|
1.573
|
|
ME1
|
malic enzyme 1, NADP(+)-dependent, cytosolic
|
|
chr17_+_79651035
|
1.527
|
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate
|
|
chr17_+_79651042
|
1.514
|
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate
|
|
chr6_-_84140758
|
1.511
|
|
ME1
|
malic enzyme 1, NADP(+)-dependent, cytosolic
|
|
chr11_+_20070609
|
1.452
|
|
NAV2
|
neuron navigator 2
|
|
chr15_+_67458492
|
1.406
|
NM_001145104
|
SMAD3
|
SMAD family member 3
|
|
chr22_-_27014037
|
1.314
|
|
CRYBB1
|
crystallin, beta B1
|
|
chr16_-_1429612
|
1.290
|
NM_001193389
NM_023076
|
UNKL
|
unkempt homolog (Drosophila)-like
|
|
chr22_-_50964856
|
1.273
|
NM_001169109
|
SCO2
|
SCO cytochrome oxidase deficient homolog 2 (yeast)
|
|
chr22_-_27013959
|
1.272
|
NM_001887
|
CRYBB1
|
crystallin, beta B1
|
|
chr17_+_30771520
|
1.265
|
|
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11
|
|
chr18_+_158480
|
1.264
|
NM_001037334
NM_005151
|
USP14
|
ubiquitin specific peptidase 14 (tRNA-guanine transglycosylase)
|
|
chr16_-_28620648
|
1.256
|
NM_177530
NM_177534
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1
|
|
chr7_+_107204402
|
1.227
|
NM_181581
|
DUS4L
|
dihydrouridine synthase 4-like (S. cerevisiae)
|
|
chr11_+_69455938
|
1.217
|
|
CCND1
|
cyclin D1
|
|
chr11_+_62649160
|
1.211
|
|
SLC3A2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2
|
|
chr16_+_30077120
|
1.192
|
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr18_+_61637262
|
1.180
|
NM_001031848
NM_002640
NM_198833
|
SERPINB8
|
serpin peptidase inhibitor, clade B (ovalbumin), member 8
|
|
chr16_+_56642477
|
1.180
|
NM_005953
|
MT2A
|
metallothionein 2A
|
|
chr17_+_79651072
|
1.177
|
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate
|
|
chr3_-_186262114
|
1.172
|
NM_017541
|
CRYGS
|
crystallin, gamma S
|
|
chr6_-_134496833
|
1.152
|
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr6_-_35888816
|
1.151
|
|
SRPK1
|
SRSF protein kinase 1
|
|
chr18_+_158586
|
1.144
|
|
USP14
|
ubiquitin specific peptidase 14 (tRNA-guanine transglycosylase)
|
|
chr6_-_84140651
|
1.140
|
|
ME1
|
malic enzyme 1, NADP(+)-dependent, cytosolic
|
|
chr16_+_30076993
|
1.119
|
NM_001243177
NM_184041
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr1_-_115238091
|
1.114
|
NM_000036
NM_001172626
|
AMPD1
|
adenosine monophosphate deaminase 1
|
|
chr1_-_32264271
|
1.091
|
|
SPOCD1
|
SPOC domain containing 1
|
|
chr6_-_35888889
|
1.077
|
|
SRPK1
|
SRSF protein kinase 1
|
|
chr1_-_6662898
|
1.060
|
NM_014851
|
KLHL21
|
kelch-like 21 (Drosophila)
|
|
chr6_-_32821592
|
1.039
|
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP)
|
|
chr8_-_125740378
|
1.035
|
|
MTSS1
|
metastasis suppressor 1
|
|
chr16_+_30077146
|
1.031
|
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr5_-_59064334
|
1.031
|
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr3_+_48507209
|
1.019
|
NM_016381
NM_033629
|
TREX1
|
three prime repair exonuclease 1
|
|
chr13_+_44453419
|
1.009
|
NM_001128303
|
LACC1
|
laccase (multicopper oxidoreductase) domain containing 1
|
|
chr10_+_114136014
|
0.977
|
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr1_-_109969044
|
0.974
|
|
PSMA5
|
proteasome (prosome, macropain) subunit, alpha type, 5
|
|
chr10_+_91061705
|
0.964
|
NM_001547
|
IFIT2
|
interferon-induced protein with tetratricopeptide repeats 2
|
|
chr2_+_85980600
|
0.964
|
NM_032827
|
ATOH8
|
atonal homolog 8 (Drosophila)
|
|
chr6_-_33548017
|
0.963
|
NM_001188
|
BAK1
|
BCL2-antagonist/killer 1
|
|
chr2_-_224467049
|
0.943
|
|
SCG2
|
secretogranin II
|
|
chr6_-_35888937
|
0.943
|
NM_003137
|
SRPK1
|
SRSF protein kinase 1
|
|
chr1_-_109968951
|
0.939
|
|
PSMA5
|
proteasome (prosome, macropain) subunit, alpha type, 5
|
|
chr7_-_100881030
|
0.930
|
NM_014343
|
CLDN15
|
claudin 15
|
|
chr16_+_83986826
|
0.928
|
NM_182981
|
OSGIN1
|
oxidative stress induced growth inhibitor 1
|
|
chr11_-_3862183
|
0.921
|
NM_001665
|
RHOG
|
ras homolog gene family, member G (rho G)
|
|
chr14_+_58711457
|
0.918
|
NM_002788
NM_152132
|
PSMA3
|
proteasome (prosome, macropain) subunit, alpha type, 3
|
|
chr22_+_22723972
|
0.918
|
|
|
|
|
chr1_-_6662681
|
0.913
|
|
KLHL21
|
kelch-like 21 (Drosophila)
|
|
chr14_-_25078818
|
0.911
|
NM_033423
|
GZMH
|
granzyme H (cathepsin G-like 2, protein h-CCPX)
|
|
chr6_-_33547833
|
0.909
|
|
BAK1
|
BCL2-antagonist/killer 1
|
|
chr14_+_58711555
|
0.900
|
|
PSMA3
|
proteasome (prosome, macropain) subunit, alpha type, 3
|
|
chr19_-_49658645
|
0.899
|
NM_002152
|
HRC
|
histidine rich calcium binding protein
|
|
chr6_-_24877453
|
0.892
|
NM_015864
|
FAM65B
|
family with sequence similarity 65, member B
|
|
chr16_+_56685810
|
0.886
|
NM_005947
|
MT1B
|
metallothionein 1B
|
|
chr1_-_162838550
|
0.871
|
NM_178550
|
C1orf110
|
chromosome 1 open reading frame 110
|
|
chr20_-_633844
|
0.866
|
NM_080725
|
SRXN1
|
sulfiredoxin 1
|
|
chr1_-_109969058
|
0.863
|
NM_001199772
NM_002790
NM_001199773
NM_001199774
|
PSMA5
|
proteasome (prosome, macropain) subunit, alpha type, 5
|
|
chr17_+_30771522
|
0.862
|
|
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11
|
|
chr22_+_21369441
|
0.857
|
NM_001159554
NM_005446
|
P2RX6
|
purinergic receptor P2X, ligand-gated ion channel, 6
|
|
chr17_+_30771532
|
0.851
|
|
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11
|
|
chr6_-_134497069
|
0.843
|
NM_001143678
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr8_+_120428551
|
0.837
|
NM_002514
|
NOV
|
nephroblastoma overexpressed gene
|
|
chr10_+_121410751
|
0.836
|
NM_004281
|
BAG3
|
BCL2-associated athanogene 3
|
|
chr1_-_32264215
|
0.832
|
|
SPOCD1
|
SPOC domain containing 1
|
|
chr16_-_28621279
|
0.832
|
NM_001055
NM_177529
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1
|
|
chr3_+_48507621
|
0.830
|
|
TREX1
|
three prime repair exonuclease 1
|
|
chr2_-_224467095
|
0.826
|
|
SCG2
|
secretogranin II
|
|
chr20_+_48807365
|
0.824
|
|
CEBPB
|
CCAAT/enhancer binding protein (C/EBP), beta
|
|
chr16_-_28607694
|
0.821
|
NM_177528
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2
|
|
chr6_-_100911550
|
0.819
|
NM_005068
|
SIM1
|
single-minded homolog 1 (Drosophila)
|
|
chr19_+_13135368
|
0.811
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr5_+_179247841
|
0.811
|
NM_003900
|
SQSTM1
|
sequestosome 1
|
|
chr9_+_103191795
|
0.806
|
NM_001198806
|
C9orf30
|
chromosome 9 open reading frame 30
|
|
chr14_+_58711599
|
0.801
|
|
PSMA3
|
proteasome (prosome, macropain) subunit, alpha type, 3
|
|
chr20_-_30060815
|
0.796
|
NM_001037500
|
DEFB124
|
defensin, beta 124
|
|
chr17_-_39280377
|
0.795
|
NM_031854
|
KRTAP4-12
|
keratin associated protein 4-12
|
|
chr10_-_71892596
|
0.795
|
NM_001198696
NM_032797
|
AIFM2
|
apoptosis-inducing factor, mitochondrion-associated, 2
|
|
chr6_+_64281910
|
0.794
|
NM_003463
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr22_-_45608232
|
0.786
|
|
KIAA0930
|
KIAA0930
|
|
chr17_+_30771494
|
0.784
|
NM_002815
|
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11
|
|
chr12_+_64845839
|
0.784
|
NM_013254
|
TBK1
|
TANK-binding kinase 1
|
|
chr11_-_3862137
|
0.780
|
|
RHOG
|
ras homolog gene family, member G (rho G)
|
|
chr2_-_233641225
|
0.775
|
NM_001172416
NM_001172417
NM_002242
|
KCNJ13
|
potassium inwardly-rectifying channel, subfamily J, member 13
|
|
chr17_-_65362635
|
0.767
|
|
PSMD12
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 12
|
|
chr22_+_23010757
|
0.765
|
|
|
|
|
chr17_+_30771485
|
0.753
|
|
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11
|
|
chr22_+_23154240
|
0.750
|
|
|
|
|
chr15_+_67418053
|
0.732
|
NM_001145102
|
SMAD3
|
SMAD family member 3
|
|
chr19_-_46285746
|
0.732
|
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr3_+_183903970
|
0.718
|
|
ABCF3
|
ATP-binding cassette, sub-family F (GCN20), member 3
|
|
chr4_+_169753105
|
0.716
|
NM_001166110
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr4_-_21950373
|
0.709
|
NM_147182
|
KCNIP4
|
Kv channel interacting protein 4
|
|
chr19_-_51568323
|
0.705
|
NM_015596
|
KLK13
|
kallikrein-related peptidase 13
|
|
chr12_-_57030083
|
0.699
|
NM_013449
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A
|
|
chr8_-_131455833
|
0.696
|
NM_001247996
NM_018482
|
ASAP1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1
|
|
chr19_+_49055421
|
0.694
|
NM_177973
|
SULT2B1
|
sulfotransferase family, cytosolic, 2B, member 1
|
|
chr11_+_69455837
|
0.693
|
NM_053056
|
CCND1
|
cyclin D1
|
|
chr3_-_172428773
|
0.693
|
NM_001146276
NM_001146277
NM_001146278
NM_020792
|
NCEH1
|
neutral cholesterol ester hydrolase 1
|
|
chr5_-_59064437
|
0.692
|
NM_001197218
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr11_+_69455978
|
0.689
|
|
CCND1
|
cyclin D1
|
|
chr3_+_183903862
|
0.688
|
NM_018358
|
ABCF3
|
ATP-binding cassette, sub-family F (GCN20), member 3
|
|
chr6_+_34206567
|
0.688
|
NM_145905
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr17_-_39623476
|
0.683
|
NM_002278
|
KRT32
|
keratin 32
|
|
chrX_-_110513750
|
0.676
|
NM_014289
|
CAPN6
|
calpain 6
|
|
chr19_+_51645556
|
0.667
|
NM_014385
NM_016543
|
SIGLEC7
|
sialic acid binding Ig-like lectin 7
|
|
chr3_-_150480917
|
0.663
|
|
SIAH2
|
seven in absentia homolog 2 (Drosophila)
|
|
chr17_-_33390676
|
0.659
|
NM_001017368
|
RFFL
|
ring finger and FYVE-like domain containing 1
|
|
chr8_-_42623928
|
0.657
|
NM_001199279
NM_004198
|
CHRNA6
|
cholinergic receptor, nicotinic, alpha 6
|
|
chr21_-_36421586
|
0.655
|
NM_001754
|
RUNX1
|
runt-related transcription factor 1
|
|
chr19_+_56347943
|
0.645
|
NM_134444
|
NLRP4
|
NLR family, pyrin domain containing 4
|
|
chr2_-_224467142
|
0.642
|
NM_003469
|
SCG2
|
secretogranin II
|
|
chr10_+_114135937
|
0.641
|
NM_016234
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr7_+_1200097
|
0.640
|
|
|
|
|
chr2_-_99871569
|
0.636
|
NM_175735
|
LYG2
|
lysozyme G-like 2
|
|
chr4_+_39699663
|
0.625
|
NM_001111112
NM_001111113
NM_005339
|
UBE2K
|
ubiquitin-conjugating enzyme E2K
|
|
chr18_+_21452981
|
0.623
|
NM_000227
NM_001127718
|
LAMA3
|
laminin, alpha 3
|
|
chr16_+_89988416
|
0.623
|
NM_001197181
|
TUBB3
|
tubulin, beta 3 class III
|
|
chr3_-_39234071
|
0.622
|
NM_194293
NM_001198621
|
XIRP1
|
xin actin-binding repeat containing 1
|
|
chr1_-_24434927
|
0.622
|
|
MYOM3
|
myomesin family, member 3
|
|
chr6_+_56819706
|
0.622
|
NM_152731
|
BEND6
|
BEN domain containing 6
|
|
chr1_+_14075917
|
0.620
|
|
PRDM2
|
PR domain containing 2, with ZNF domain
|
|
chr17_+_46985720
|
0.614
|
NM_023079
|
UBE2Z
|
ubiquitin-conjugating enzyme E2Z
|
|
chr7_-_1199813
|
0.614
|
|
ZFAND2A
|
zinc finger, AN1-type domain 2A
|
|
chr3_-_150480963
|
0.612
|
NM_005067
|
SIAH2
|
seven in absentia homolog 2 (Drosophila)
|
|
chr18_+_21594349
|
0.611
|
NM_001135993
NM_001243425
|
TTC39C
|
tetratricopeptide repeat domain 39C
|
|
chr10_-_71892514
|
0.604
|
|
AIFM2
|
apoptosis-inducing factor, mitochondrion-associated, 2
|
|
chr6_+_160542862
|
0.590
|
NM_003057
NM_153187
|
SLC22A1
|
solute carrier family 22 (organic cation transporter), member 1
|
|
chr2_+_69201704
|
0.586
|
NM_019617
|
GKN1
|
gastrokine 1
|
|
chr10_+_114135965
|
0.580
|
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr19_+_36359346
|
0.576
|
NM_001024807
NM_005166
|
APLP1
|
amyloid beta (A4) precursor-like protein 1
|
|
chr15_-_42448769
|
0.574
|
NM_213600
|
PLA2G4F
|
phospholipase A2, group IVF
|
|
chr14_+_23352430
|
0.573
|
NM_173527
|
REM2
|
RAS (RAD and GEM)-like GTP binding 2
|
|
chr5_-_172756505
|
0.572
|
NM_003714
|
STC2
|
stanniocalcin 2
|
|
chr17_+_73717515
|
0.563
|
NM_000213
NM_001005731
|
ITGB4
|
integrin, beta 4
|
|
chr11_-_9025531
|
0.561
|
NM_020645
|
NRIP3
|
nuclear receptor interacting protein 3
|
|
chr1_+_179262936
|
0.558
|
|
SOAT1
|
sterol O-acyltransferase 1
|
|
chr19_+_38865184
|
0.550
|
NM_002812
|
PSMD8
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 8
|
|
chr3_-_79068542
|
0.548
|
NM_001145845
NM_133631
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr10_+_121410891
|
0.542
|
|
BAG3
|
BCL2-associated athanogene 3
|
|
chr17_+_48610077
|
0.537
|
|
EPN3
|
epsin 3
|
|
chr3_+_11178778
|
0.535
|
NM_001098213
|
HRH1
|
histamine receptor H1
|
|
chr12_+_69202796
|
0.534
|
|
MDM2
|
Mdm2 p53 binding protein homolog (mouse)
|
|
chr21_-_36260086
|
0.534
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr9_-_140083010
|
0.526
|
|
ANAPC2
|
anaphase promoting complex subunit 2
|
|
chr15_+_40733302
|
0.521
|
NM_014952
|
BAHD1
|
bromo adjacent homology domain containing 1
|
|
chr20_+_48807119
|
0.518
|
NM_005194
|
CEBPB
|
CCAAT/enhancer binding protein (C/EBP), beta
|
|
chr22_-_40859423
|
0.516
|
|
MKL1
|
megakaryoblastic leukemia (translocation) 1
|
|
chr9_-_34458477
|
0.513
|
NM_001184940
NM_001184941
NM_001184942
NM_001184943
NM_001184945
NM_147202
|
C9orf25
|
chromosome 9 open reading frame 25
|
|
chr22_+_22676814
|
0.511
|
|
|
|
|
chr1_+_233749749
|
0.506
|
NM_002245
|
KCNK1
|
potassium channel, subfamily K, member 1
|
|
chr10_+_121411086
|
0.505
|
|
BAG3
|
BCL2-associated athanogene 3
|
|
chr1_+_14075931
|
0.504
|
|
PRDM2
|
PR domain containing 2, with ZNF domain
|
|
chr7_-_102985068
|
0.503
|
NM_001129887
NM_014377
|
DNAJC2
|
DnaJ (Hsp40) homolog, subfamily C, member 2
|
|
chr4_-_21699317
|
0.502
|
NM_001035003
|
KCNIP4
|
Kv channel interacting protein 4
|
|
chr3_-_53916087
|
0.501
|
|
ACTR8
|
ARP8 actin-related protein 8 homolog (yeast)
|
|
chr8_-_26724748
|
0.493
|
|
ADRA1A
|
adrenergic, alpha-1A-, receptor
|
|
chr10_+_121411042
|
0.479
|
|
BAG3
|
BCL2-associated athanogene 3
|
|
chr5_+_179247903
|
0.474
|
|
SQSTM1
|
sequestosome 1
|
|
chr5_-_35938736
|
0.472
|
NM_001042625
NM_144647
|
CAPSL
|
calcyphosine-like
|
|
chr20_+_24929904
|
0.472
|
|
CST7
|
cystatin F (leukocystatin)
|
|
chr19_-_8212367
|
0.469
|
NM_032447
|
FBN3
|
fibrillin 3
|
|
chr16_+_74330730
|
0.468
|
|
PSMD7
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 7
|
|
chr4_-_87281374
|
0.464
|
NM_138980
NM_138982
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr2_+_105471745
|
0.464
|
NM_006236
|
POU3F3
|
POU class 3 homeobox 3
|
|
chr9_-_113018869
|
0.461
|
NM_001244938
NM_003329
|
TXN
|
thioredoxin
|
|
chr17_-_48264039
|
0.461
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr5_-_175843360
|
0.459
|
|
CLTB
|
clathrin, light chain B
|
|
chr7_+_129932973
|
0.457
|
NM_001163446
NM_016352
|
CPA4
|
carboxypeptidase A4
|
|
chr7_-_75988315
|
0.455
|
NM_012479
|
YWHAG
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide
|
|
chrX_-_70326637
|
0.446
|
NM_001025265
|
CXorf65
|
chromosome X open reading frame 65
|
|
chr12_+_7023490
|
0.445
|
NM_001975
|
ENO2
|
enolase 2 (gamma, neuronal)
|
|
chr3_-_170587947
|
0.444
|
NM_001099645
|
RPL22L1
|
ribosomal protein L22-like 1
|
|
chr1_-_150208441
|
0.441
|
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E
|