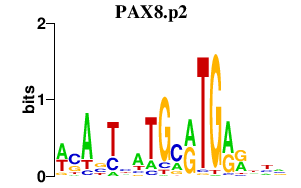
|
chr13_+_97928196
|
0.189
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr10_+_85933553
|
0.177
|
NM_207373
|
C10orf99
|
chromosome 10 open reading frame 99
|
|
chr4_+_36283243
|
0.157
|
NM_001136536
|
DTHD1
|
death domain containing 1
|
|
chr8_+_39792473
|
0.131
|
NM_194294
|
IDO2
|
indoleamine 2,3-dioxygenase 2
|
|
chr3_-_71542628
|
0.123
|
|
FOXP1
|
forkhead box P1
|
|
chr6_-_31107110
|
0.116
|
NM_014069
|
PSORS1C2
|
psoriasis susceptibility 1 candidate 2
|
|
chrX_-_100307091
|
0.097
|
NM_001167970
NM_001167971
NM_001167972
NM_024917
|
TRMT2B
|
TRM2 tRNA methyltransferase 2 homolog B (S. cerevisiae)
|
|
chrX_-_100307042
|
0.095
|
|
TRMT2B
|
TRM2 tRNA methyltransferase 2 homolog B (S. cerevisiae)
|
|
chr17_-_63009302
|
0.087
|
|
GNA13
|
guanine nucleotide binding protein (G protein), alpha 13
|
|
chr5_-_94224601
|
0.072
|
|
MCTP1
|
multiple C2 domains, transmembrane 1
|
|
chr7_+_129984629
|
0.061
|
NM_001127441
NM_001127442
NM_080385
|
CPA5
|
carboxypeptidase A5
|
|
chr3_-_9290592
|
0.061
|
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3
|
|
chr1_-_27952591
|
0.053
|
NM_001042747
|
FGR
|
Gardner-Rasheed feline sarcoma viral (v-fgr) oncogene homolog
|
|
chr1_-_93399531
|
0.052
|
|
|
|
|
chr18_+_59157774
|
0.035
|
NM_031891
|
CDH20
|
cadherin 20, type 2
|
|
chr9_+_124048869
|
0.028
|
|
GSN
|
gelsolin
|
|
chr1_+_111772332
|
0.027
|
NM_001025199
|
CHI3L2
|
chitinase 3-like 2
|
|
chrX_+_37892786
|
0.023
|
NM_001163334
|
SYTL5
|
synaptotagmin-like 5
|
|
chr3_-_189840225
|
0.020
|
NM_001134418
|
LEPREL1
|
leprecan-like 1
|
|
chr17_-_39646115
|
0.017
|
NM_003771
|
KRT36
|
keratin 36
|
|
chr19_+_9361605
|
0.016
|
NM_001079935
|
OR7E24
|
olfactory receptor, family 7, subfamily E, member 24
|
|
chr6_+_41196061
|
0.014
|
NM_198153
|
TREML4
|
triggering receptor expressed on myeloid cells-like 4
|
|
chr7_+_110731061
|
0.012
|
NM_001099658
NM_001099660
NM_018334
|
LRRN3
|
leucine rich repeat neuronal 3
|
|
chr10_+_103911932
|
0.011
|
NM_004741
|
NOLC1
|
nucleolar and coiled-body phosphoprotein 1
|
|
chr9_+_125376947
|
0.011
|
NM_012364
|
OR1Q1
|
olfactory receptor, family 1, subfamily Q, member 1
|
|
chr19_-_51537996
|
0.009
|
NM_019598
NM_145894
NM_145895
|
KLK12
|
kallikrein-related peptidase 12
|
|
chr3_-_157213138
|
0.007
|
NM_001167917
|
VEPH1
|
ventricular zone expressed PH domain homolog 1 (zebrafish)
|
|
chr3_-_9291251
|
0.007
|
NM_001033117
NM_014850
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3
|
|
chr12_-_9360960
|
0.007
|
NM_002864
|
PZP
|
pregnancy-zone protein
|
|
chr9_-_4666665
|
0.006
|
NM_001039395
|
C9orf68
|
chromosome 9 open reading frame 68
|
|
chr12_-_102872329
|
0.005
|
NM_001111284
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr2_-_179672056
|
0.005
|
NM_003319
NM_133378
NM_133379
NM_133432
NM_133437
|
TTN
|
titin
|
|
chr4_-_10686314
|
0.005
|
NM_052964
|
CLNK
|
cytokine-dependent hematopoietic cell linker
|
|
chr17_+_7461591
|
0.003
|
NM_001198622
NM_001198623
NM_001198624
NM_003808
NM_172087
NM_172088
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13
|
|
chr4_-_69434215
|
0.003
|
NM_001077
|
UGT2B15
UGT2B17
|
UDP glucuronosyltransferase 2 family, polypeptide B15
UDP glucuronosyltransferase 2 family, polypeptide B17
|
|
chrX_-_153141398
|
0.001
|
NM_000425
NM_001143963
NM_024003
|
L1CAM
|
L1 cell adhesion molecule
|