|
chr2_+_152214105
|
1.905
|
NM_007115
|
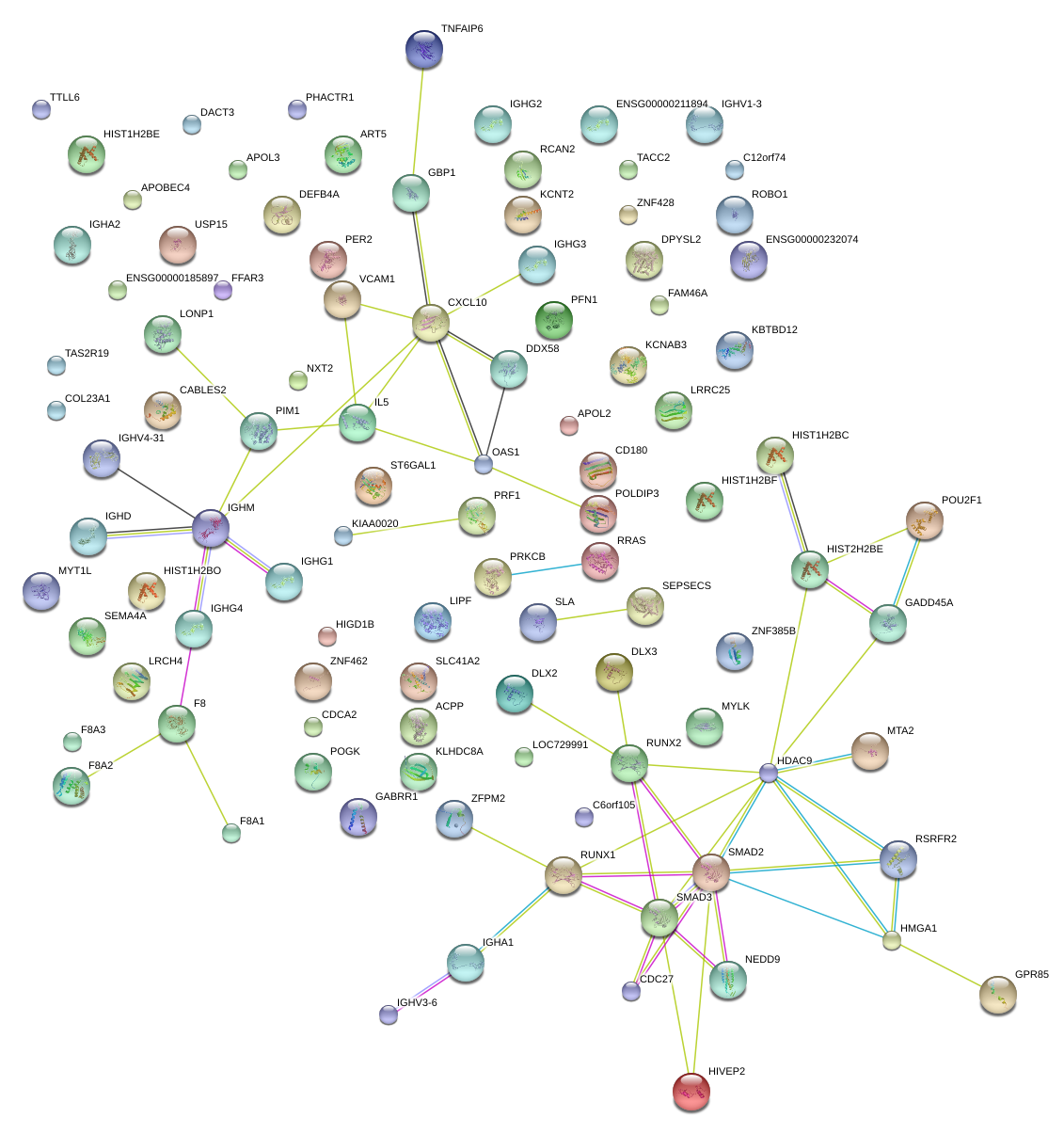
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6
|
|
chr12_+_113344738
|
1.462
|
NM_001032409
NM_002534
NM_016816
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa
|
|
chr6_-_11779279
|
1.290
|
NM_001143948
NM_032744
|
C6orf105
|
chromosome 6 open reading frame 105
|
|
chr3_-_123603144
|
1.272
|
NM_053025
NM_053026
NM_053027
NM_053028
|
MYLK
|
myosin light chain kinase
|
|
chr15_+_67458492
|
1.071
|
NM_001145104
|
SMAD3
|
SMAD family member 3
|
|
chr1_+_68150858
|
1.022
|
NM_001199741
NM_001199742
NM_001924
|
GADD45A
|
growth arrest and DNA-damage-inducible, alpha
|
|
chr1_+_68151092
|
1.011
|
|
GADD45A
|
growth arrest and DNA-damage-inducible, alpha
|
|
chr4_-_76944585
|
0.966
|
NM_001565
|
CXCL10
|
chemokine (C-X-C motif) ligand 10
|
|
chr12_+_113344811
|
0.883
|
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa
|
|
chr12_-_105322056
|
0.883
|
NM_032148
|
SLC41A2
|
solute carrier family 41, member 2
|
|
chr3_-_79817058
|
0.877
|
NM_002941
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr21_-_36260972
|
0.849
|
NM_001001890
NM_001122607
|
RUNX1
|
runt-related transcription factor 1
|
|
chr8_+_26435590
|
0.833
|
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr8_+_106331146
|
0.803
|
NM_012082
|
ZFPM2
|
zinc finger protein, multitype 2
|
|
chr6_-_82462374
|
0.777
|
NM_017633
|
FAM46A
|
family with sequence similarity 46, member A
|
|
chr9_+_109686436
|
0.772
|
|
ZNF462
|
zinc finger protein 462
|
|
chr1_-_205313309
|
0.746
|
|
KLHDC8A
|
kelch domain containing 8A
|
|
chr11_-_62369213
|
0.746
|
|
MTA2
|
metastasis associated 1 family, member 2
|
|
chr6_-_143266283
|
0.740
|
NM_006734
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2
|
|
chr14_-_106692107
|
0.739
|
|
IGHV4-31
IGHA1
IGHD
IGHG1
IGHG3
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
|
|
chr11_-_62369097
|
0.734
|
|
MTA2
|
metastasis associated 1 family, member 2
|
|
chr9_-_32526164
|
0.733
|
NM_014314
|
DDX58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58
|
|
chr6_+_37137882
|
0.707
|
NM_001243186
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr11_-_62369250
|
0.705
|
NM_004739
|
MTA2
|
metastasis associated 1 family, member 2
|
|
chr19_+_35849487
|
0.652
|
NM_005304
|
FFAR3
|
free fatty acid receptor 3
|
|
chr7_+_18535351
|
0.646
|
NM_001204145
NM_001204146
NM_014707
NM_178423
|
HDAC9
|
histone deacetylase 9
|
|
chr6_-_89927495
|
0.637
|
NM_002042
|
GABRR1
|
gamma-aminobutyric acid (GABA) receptor, rho 1
|
|
chr5_-_66492574
|
0.621
|
NM_005582
|
CD180
|
CD180 molecule
|
|
chr12_-_11175169
|
0.608
|
NM_176888
|
TAS2R19
|
taste receptor, type 2, member 19
|
|
chr11_-_3663484
|
0.599
|
NM_053017
NM_001079536
|
ART5
|
ADP-ribosyltransferase 5
|
|
chr7_-_100183722
|
0.595
|
|
LRCH4
|
leucine-rich repeats and calponin homology (CH) domain containing 4
|
|
chr1_-_196577438
|
0.574
|
NM_198503
|
KCNT2
|
potassium channel, subfamily T, member 2
|
|
chr1_-_89530890
|
0.563
|
NM_002053
|
GBP1
|
guanylate binding protein 1, interferon-inducible
|
|
chr1_-_183622447
|
0.549
|
NM_203454
|
APOBEC4
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 4 (putative)
|
|
chr8_-_7274353
|
0.546
|
NM_001205266
|
DEFB4B
|
defensin, beta 4B
|
|
chr14_-_107218786
|
0.527
|
|
IGHA1
IGHG1
IGHM
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant mu
|
|
chr1_+_156119734
|
0.503
|
NM_001193301
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr6_+_34204928
|
0.502
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr16_+_23847321
|
0.498
|
|
PRKCB
|
protein kinase C, beta
|
|
chr3_+_127641901
|
0.483
|
NM_207335
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12
|
|
chr7_-_112727832
|
0.479
|
NM_001146265
NM_018970
|
GPR85
|
G protein-coupled receptor 85
|
|
chr22_-_36556781
|
0.467
|
NM_145640
|
APOL3
|
apolipoprotein L, 3
|
|
chrX_+_154611748
|
0.464
|
NM_001007523
NM_001007524
NM_012151
|
F8A2
F8A3
F8A1
|
coagulation factor VIII-associated 2
coagulation factor VIII-associated 3
coagulation factor VIII-associated 1
|
|
chr1_+_101185195
|
0.450
|
NM_001078
NM_001199834
NM_080682
|
VCAM1
|
vascular cell adhesion molecule 1
|
|
chr16_-_55909208
|
0.427
|
NM_001143685
NM_145024
|
CES5A
|
carboxylesterase 5A
|
|
chr6_+_34204690
|
0.426
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr6_-_11232900
|
0.425
|
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr6_+_31588460
|
0.416
|
|
PRRC2A
|
proline-rich coiled-coil 2A
|
|
chr19_-_19281090
|
0.413
|
NM_001145785
|
MEF2B
|
myocyte enhancer factor 2B
|
|
chr7_-_112727778
|
0.412
|
|
GPR85
|
G protein-coupled receptor 85
|
|
chrX_-_154688260
|
0.412
|
NM_001007523
NM_001007524
NM_012151
|
F8A2
F8A3
F8A1
|
coagulation factor VIII-associated 2
coagulation factor VIII-associated 3
coagulation factor VIII-associated 1
|
|
chr2_-_239197091
|
0.407
|
NM_022817
|
PER2
|
period homolog 2 (Drosophila)
|
|
chr10_+_123923104
|
0.407
|
NM_006997
NM_206860
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr8_+_7752198
|
0.402
|
NM_004942
|
DEFB4A
|
defensin, beta 4A
|
|
chr3_+_132036276
|
0.402
|
|
ACPP
|
acid phosphatase, prostate
|
|
chr19_-_19281062
|
0.396
|
|
MEF2B
|
myocyte enhancer factor 2B
|
|
chr19_-_50143350
|
0.394
|
NM_006270
|
RRAS
|
related RAS viral (r-ras) oncogene homolog
|
|
chr19_-_18508407
|
0.392
|
NM_145256
|
LRRC25
|
leucine rich repeat containing 25
|
|
chr10_+_123922940
|
0.392
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr7_-_112727799
|
0.388
|
|
GPR85
|
G protein-coupled receptor 85
|
|
chr6_+_31588524
|
0.387
|
|
PRRC2A
|
proline-rich coiled-coil 2A
|
|
chr8_+_25316512
|
0.384
|
NM_152562
|
CDCA2
|
cell division cycle associated 2
|
|
chr6_+_34204576
|
0.381
|
NM_002131
NM_145899
NM_145903
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr1_+_167298280
|
0.373
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr5_-_178017555
|
0.369
|
NM_173465
|
COL23A1
|
collagen, type XXIII, alpha 1
|
|
chr6_-_46293628
|
0.369
|
NM_005822
|
RCAN2
|
regulator of calcineurin 2
|
|
chr12_+_62654120
|
0.363
|
NM_001252078
NM_001252079
NM_006313
|
USP15
|
ubiquitin specific peptidase 15
|
|
chr17_+_42925278
|
0.363
|
NM_016438
|
HIGD1B
|
HIG1 hypoxia inducible domain family, member 1B
|
|
chr19_-_44123732
|
0.362
|
|
ZNF428
|
zinc finger protein 428
|
|
chr14_-_107013122
|
0.362
|
|
IGHV4-31
IGHG1
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr2_-_180427314
|
0.359
|
NM_001113398
|
ZNF385B
|
zinc finger protein 385B
|
|
chr22_-_43010892
|
0.358
|
|
POLDIP3
|
polymerase (DNA-directed), delta interacting protein 3
|
|
chr9_-_2844083
|
0.358
|
|
KIAA0020
|
KIAA0020
|
|
chr2_-_2334887
|
0.355
|
NM_015025
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr17_-_46894380
|
0.353
|
NM_001130918
|
TTLL6
|
tubulin tyrosine ligase-like family, member 6
|
|
chr22_-_43010942
|
0.352
|
NM_032311
NM_178136
|
POLDIP3
|
polymerase (DNA-directed), delta interacting protein 3
|
|
chr9_-_2844098
|
0.349
|
NM_014878
|
KIAA0020
|
KIAA0020
|
|
chr6_+_34204663
|
0.347
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr6_+_31588444
|
0.345
|
NM_004638
NM_080686
|
PRRC2A
|
proline-rich coiled-coil 2A
|
|
chr17_-_7832752
|
0.344
|
NM_004732
|
KCNAB3
|
potassium voltage-gated channel, shaker-related subfamily, beta member 3
|
|
chr6_+_12717036
|
0.343
|
NM_001242648
NM_030948
|
PHACTR1
|
phosphatase and actin regulator 1
|
|
chr10_+_90424145
|
0.342
|
NM_001198828
NM_001198829
NM_001198830
NM_004190
|
LIPF
|
lipase, gastric
|
|
chr22_+_22786283
|
0.341
|
|
|
|
|
chr19_-_47164289
|
0.339
|
NM_145056
|
DACT3
|
dapper, antagonist of beta-catenin, homolog 3 (Xenopus laevis)
|
|
chr6_+_31588523
|
0.339
|
|
PRRC2A
|
proline-rich coiled-coil 2A
|
|
chr3_+_186648273
|
0.336
|
NM_173216
NM_173217
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr5_-_131879172
|
0.335
|
NM_000879
|
IL5
|
interleukin 5 (colony-stimulating factor, eosinophil)
|
|
chr20_-_60982334
|
0.330
|
NM_031215
|
CABLES2
|
Cdk5 and Abl enzyme substrate 2
|
|
chr12_+_93096618
|
0.323
|
NM_001037671
NM_001178097
|
C12orf74
|
chromosome 12 open reading frame 74
|
|
chr22_-_43010837
|
0.321
|
|
POLDIP3
|
polymerase (DNA-directed), delta interacting protein 3
|
|
chr6_+_26183957
|
0.320
|
NM_003523
|
HIST1H2BE
|
histone cluster 1, H2be
|
|
chr1_+_166808721
|
0.316
|
NM_017542
|
POGK
|
pogo transposable element with KRAB domain
|
|
chr17_-_4851797
|
0.314
|
NM_005022
|
PFN1
|
profilin 1
|
|
chr8_+_25316714
|
0.311
|
|
CDCA2
|
cell division cycle associated 2
|
|
chrX_+_108779009
|
0.310
|
NM_018698
|
NXT2
|
nuclear transport factor 2-like export factor 2
|
|
chr8_-_134115309
|
0.308
|
NM_001045556
NM_001045557
|
SLA
|
Src-like-adaptor
|
|
chr17_-_48072566
|
0.308
|
NM_005220
|
DLX3
|
distal-less homeobox 3
|
|
chr11_-_568419
|
0.306
|
|
MIR210HG
|
MIR210 host gene (non-protein coding)
|
|
chr14_+_23352430
|
0.305
|
NM_173527
|
REM2
|
RAS (RAD and GEM)-like GTP binding 2
|
|
chr2_+_54785453
|
0.302
|
NM_178313
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1
|
|
chr17_+_72983723
|
0.299
|
NM_014603
|
CDR2L
|
cerebellar degeneration-related protein 2-like
|
|
chr8_+_123793988
|
0.299
|
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr10_+_97759847
|
0.299
|
NM_001001732
NM_001159747
|
CC2D2B
|
coiled-coil and C2 domain containing 2B
|
|
chr3_-_46068978
|
0.298
|
NM_001024644
NM_005283
|
XCR1
|
chemokine (C motif) receptor 1
|
|
chr1_-_103496768
|
0.297
|
|
COL11A1
|
collagen, type XI, alpha 1
|
|
chr6_+_34204647
|
0.296
|
NM_145901
NM_145902
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr6_-_53013581
|
0.295
|
NM_003643
|
GCM1
|
glial cells missing homolog 1 (Drosophila)
|
|
chr7_-_122339186
|
0.295
|
NM_139175
|
RNF133
|
ring finger protein 133
|
|
chr19_-_44124013
|
0.293
|
NM_182498
|
ZNF428
|
zinc finger protein 428
|
|
chr14_-_106725232
|
0.293
|
|
ZCWPW2
IGHV3-23
IGKV3-20
IGK@
|
zinc finger, CW type with PWWP domain 2
immunoglobulin heavy variable 3-23
immunoglobulin kappa variable 3-20
immunoglobulin kappa locus
|
|
chr3_-_192445344
|
0.290
|
NM_004113
|
FGF12
|
fibroblast growth factor 12
|
|
chr14_-_68283293
|
0.288
|
NM_015346
|
ZFYVE26
|
zinc finger, FYVE domain containing 26
|
|
chr17_+_72732958
|
0.287
|
NM_001163989
|
RAB37
|
RAB37, member RAS oncogene family
|
|
chr6_-_27114577
|
0.284
|
NM_080593
|
HIST1H2BK
|
histone cluster 1, H2bk
|
|
chr6_-_168476531
|
0.284
|
NM_001122841
|
FRMD1
|
FERM domain containing 1
|
|
chr8_+_123793894
|
0.283
|
NM_014943
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr22_-_43010894
|
0.279
|
|
POLDIP3
|
polymerase (DNA-directed), delta interacting protein 3
|
|
chr17_-_36105005
|
0.275
|
NM_000458
NM_001165923
|
HNF1B
|
HNF1 homeobox B
|
|
chr1_+_149822627
|
0.273
|
NM_001040874
NM_003516
|
HIST2H2AA4
HIST2H2AA3
|
histone cluster 2, H2aa4
histone cluster 2, H2aa3
|
|
chr19_+_19144520
|
0.266
|
|
ARMC6
|
armadillo repeat containing 6
|
|
chr15_-_89089792
|
0.265
|
NM_001144074
NM_017996
|
DET1
|
de-etiolated homolog 1 (Arabidopsis)
|
|
chr1_-_149814264
|
0.265
|
NM_001040874
NM_003516
|
HIST2H2AA4
HIST2H2AA3
|
histone cluster 2, H2aa4
histone cluster 2, H2aa3
|
|
chr9_-_127533397
|
0.258
|
NM_001489
NM_033334
|
NR6A1
|
nuclear receptor subfamily 6, group A, member 1
|
|
chr11_+_537521
|
0.257
|
NM_198075
|
LRRC56
|
leucine rich repeat containing 56
|
|
chr2_-_30144431
|
0.256
|
NM_004304
|
ALK
|
anaplastic lymphoma receptor tyrosine kinase
|
|
chr6_+_10585978
|
0.255
|
NM_145655
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group)
|
|
chr3_+_119316694
|
0.251
|
NM_001206960
NM_001206961
NM_015900
|
PLA1A
|
phospholipase A1 member A
|
|
chr16_-_88752788
|
0.250
|
|
SNAI3
|
snail homolog 3 (Drosophila)
|
|
chr4_-_110223798
|
0.248
|
NM_032518
NM_198721
|
COL25A1
|
collagen, type XXV, alpha 1
|
|
chr7_-_27239724
|
0.244
|
NM_000522
|
HOXA13
|
homeobox A13
|
|
chr19_+_19144617
|
0.243
|
|
ARMC6
|
armadillo repeat containing 6
|
|
chr5_-_2751768
|
0.241
|
NM_001134222
NM_033267
|
IRX2
|
iroquois homeobox 2
|
|
chr22_-_24096551
|
0.238
|
NM_013378
|
VPREB3
|
pre-B lymphocyte 3
|
|
chr5_-_36301999
|
0.237
|
NM_001161429
NM_145000
|
RANBP3L
|
RAN binding protein 3-like
|
|
chr1_+_201979747
|
0.236
|
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr22_+_22385340
|
0.234
|
|
|
|
|
chr1_+_218518675
|
0.234
|
NM_001135599
NM_003238
|
TGFB2
|
transforming growth factor, beta 2
|
|
chr6_-_11232901
|
0.231
|
NM_006403
NM_182966
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr22_+_24033047
|
0.231
|
NM_153615
|
RGL4
|
ral guanine nucleotide dissociation stimulator-like 4
|
|
chr1_-_2457988
|
0.229
|
NM_018216
|
PANK4
|
pantothenate kinase 4
|
|
chr16_+_30107742
|
0.227
|
|
|
|
|
chr3_-_164914448
|
0.226
|
NM_014926
|
SLITRK3
|
SLIT and NTRK-like family, member 3
|
|
chr7_+_16566504
|
0.225
|
NM_001195280
|
LRRC72
|
leucine rich repeat containing 72
|
|
chr10_-_62332666
|
0.223
|
NM_001204404
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr6_-_52668599
|
0.221
|
NM_145740
|
GSTA1
|
glutathione S-transferase alpha 1
|
|
chr6_+_27114860
|
0.221
|
NM_080596
|
HIST1H2AH
|
histone cluster 1, H2ah
|
|
chr14_-_106610338
|
0.220
|
|
IGHV4-31
IGHG1
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr15_-_83240499
|
0.219
|
NM_001079533
NM_001079534
NM_001079535
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr11_-_114430579
|
0.218
|
NM_152315
|
FAM55A
|
family with sequence similarity 55, member A
|
|
chr18_+_32290195
|
0.218
|
NM_001198941
NM_032978
|
DTNA
|
dystrobrevin, alpha
|
|
chr6_-_31560497
|
0.218
|
NM_001145466
NM_001145467
NM_147130
|
NCR3
|
natural cytotoxicity triggering receptor 3
|
|
chr3_+_63638343
|
0.217
|
NM_001080537
|
SNTN
|
sentan, cilia apical structure protein
|
|
chr16_+_23847271
|
0.215
|
NM_002738
NM_212535
|
PRKCB
|
protein kinase C, beta
|
|
chr6_+_34205029
|
0.215
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr7_-_41742666
|
0.215
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr6_-_26124130
|
0.215
|
NM_003526
|
HIST1H2BG
HIST1H2BC
|
histone cluster 1, H2bg
histone cluster 1, H2bc
|
|
chr15_-_66648991
|
0.211
|
NM_017858
|
TIPIN
|
TIMELESS interacting protein
|
|
chr2_-_239148545
|
0.209
|
|
HES6
|
hairy and enhancer of split 6 (Drosophila)
|
|
chrX_+_69674914
|
0.209
|
NM_001166278
|
DLG3
|
discs, large homolog 3 (Drosophila)
|
|
chr19_-_47291594
|
0.207
|
NM_005628
|
SLC1A5
|
solute carrier family 1 (neutral amino acid transporter), member 5
|
|
chr5_-_150466696
|
0.206
|
NM_001252391
NM_001252392
|
TNIP1
|
TNFAIP3 interacting protein 1
|
|
chr17_-_73127787
|
0.206
|
NM_001252377
NM_014595
|
NT5C
|
5', 3'-nucleotidase, cytosolic
|
|
chr18_+_77866914
|
0.205
|
NM_014913
|
ADNP2
|
ADNP homeobox 2
|
|
chr1_+_203444955
|
0.204
|
|
PRELP
|
proline/arginine-rich end leucine-rich repeat protein
|
|
chr22_+_21986968
|
0.202
|
NM_152612
|
CCDC116
|
coiled-coil domain containing 116
|
|
chr11_-_34379526
|
0.201
|
NM_145804
|
ABTB2
|
ankyrin repeat and BTB (POZ) domain containing 2
|
|
chr8_+_123794043
|
0.199
|
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr11_+_69455978
|
0.197
|
|
CCND1
|
cyclin D1
|
|
chr2_-_239148626
|
0.196
|
NM_001142853
NM_018645
|
HES6
|
hairy and enhancer of split 6 (Drosophila)
|
|
chr18_+_32073245
|
0.194
|
NM_001198945
NM_001392
NM_032975
NM_032979
|
DTNA
|
dystrobrevin, alpha
|
|
chr2_-_178753464
|
0.194
|
NM_001077196
|
PDE11A
|
phosphodiesterase 11A
|
|
chr8_-_25315941
|
0.193
|
NM_017634
|
KCTD9
|
potassium channel tetramerisation domain containing 9
|
|
chr17_-_16875388
|
0.192
|
NM_012452
|
TNFRSF13B
|
tumor necrosis factor receptor superfamily, member 13B
|
|
chr14_-_107048697
|
0.192
|
|
IGHG1
|
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr4_+_184020343
|
0.191
|
NM_024949
|
WWC2
|
WW and C2 domain containing 2
|
|
chr5_+_76114832
|
0.189
|
NM_005242
|
F2RL1
|
coagulation factor II (thrombin) receptor-like 1
|
|
chr20_+_62795773
|
0.184
|
|
MYT1
|
myelin transcription factor 1
|
|
chr20_+_34556528
|
0.182
|
NM_001207076
NM_080834
|
C20orf152
|
chromosome 20 open reading frame 152
|
|
chr2_-_158182154
|
0.182
|
NM_020711
|
ERMN
|
ermin, ERM-like protein
|
|
chr1_-_68962679
|
0.180
|
|
DEPDC1
|
DEP domain containing 1
|
|
chr1_-_186649421
|
0.180
|
NM_000963
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr4_-_155312448
|
0.175
|
NM_017639
|
DCHS2
|
dachsous 2 (Drosophila)
|
|
chr6_-_10415438
|
0.175
|
NM_003220
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr10_-_98031154
|
0.173
|
NM_001114094
NM_013314
|
BLNK
|
B-cell linker
|
|
chr17_+_57642885
|
0.173
|
NM_001166301
NM_024612
|
DHX40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40
|
|
chr5_+_173472737
|
0.170
|
|
HMP19
|
HMP19 protein
|
|
chr22_+_22764094
|
0.169
|
|
|
|
|
chr1_+_159409511
|
0.166
|
NM_012351
|
OR10J1
|
olfactory receptor, family 10, subfamily J, member 1
|
|
chr6_-_11232883
|
0.164
|
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr5_-_58571916
|
0.162
|
NM_001197220
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr3_-_71632741
|
0.162
|
NM_001244808
NM_001012505
NM_001244810
NM_032682
|
FOXP1
|
forkhead box P1
|
|
chr13_-_70682458
|
0.161
|
NM_020866
|
KLHL1
|
kelch-like 1 (Drosophila)
|
|
chr8_-_25902639
|
0.161
|
NM_022659
|
EBF2
|
early B-cell factor 2
|
|
chr2_-_207082626
|
0.159
|
NM_001098199
NM_005279
|
GPR1
|
G protein-coupled receptor 1
|
|
chr8_-_25315791
|
0.159
|
|
KCTD9
|
potassium channel tetramerisation domain containing 9
|
|
chr3_+_63428752
|
0.159
|
NM_144642
|
SYNPR
|
synaptoporin
|
|
chr14_-_107113943
|
0.159
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr6_+_43139190
|
0.158
|
|
SRF
|
serum response factor (c-fos serum response element-binding transcription factor)
|
|
chrX_-_84634742
|
0.157
|
NM_024921
|
POF1B
|
premature ovarian failure, 1B
|
|
chr11_-_5345530
|
0.157
|
NM_033180
|
OR51B2
|
olfactory receptor, family 51, subfamily B, member 2
|