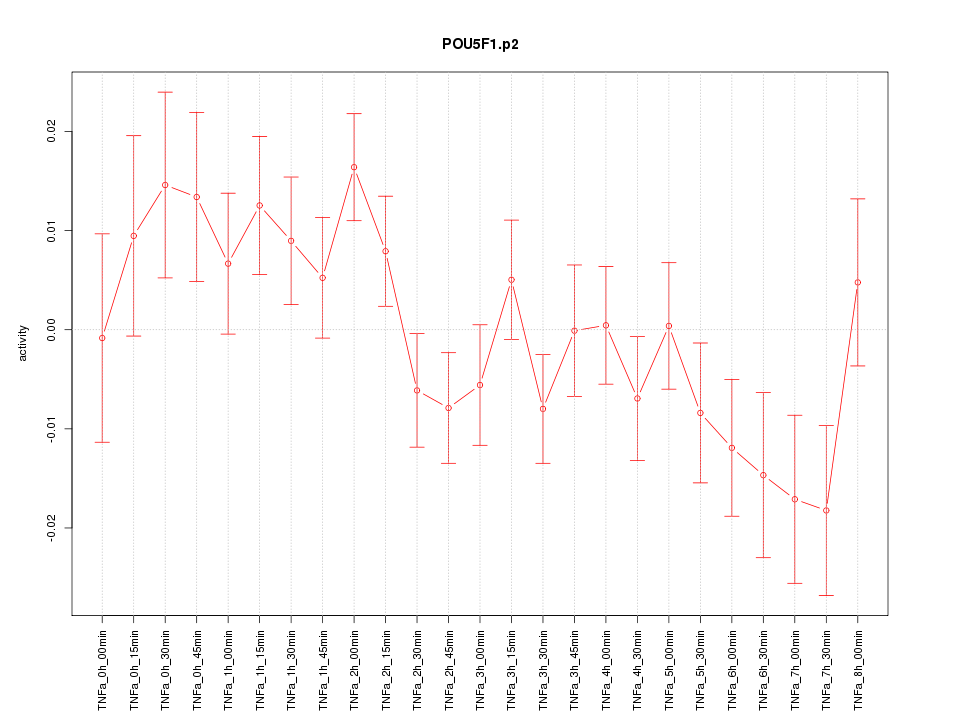
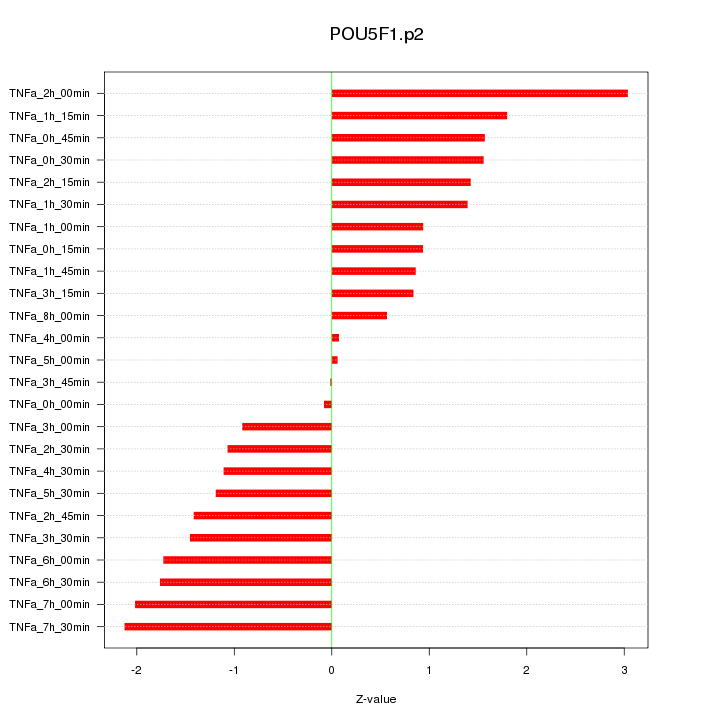
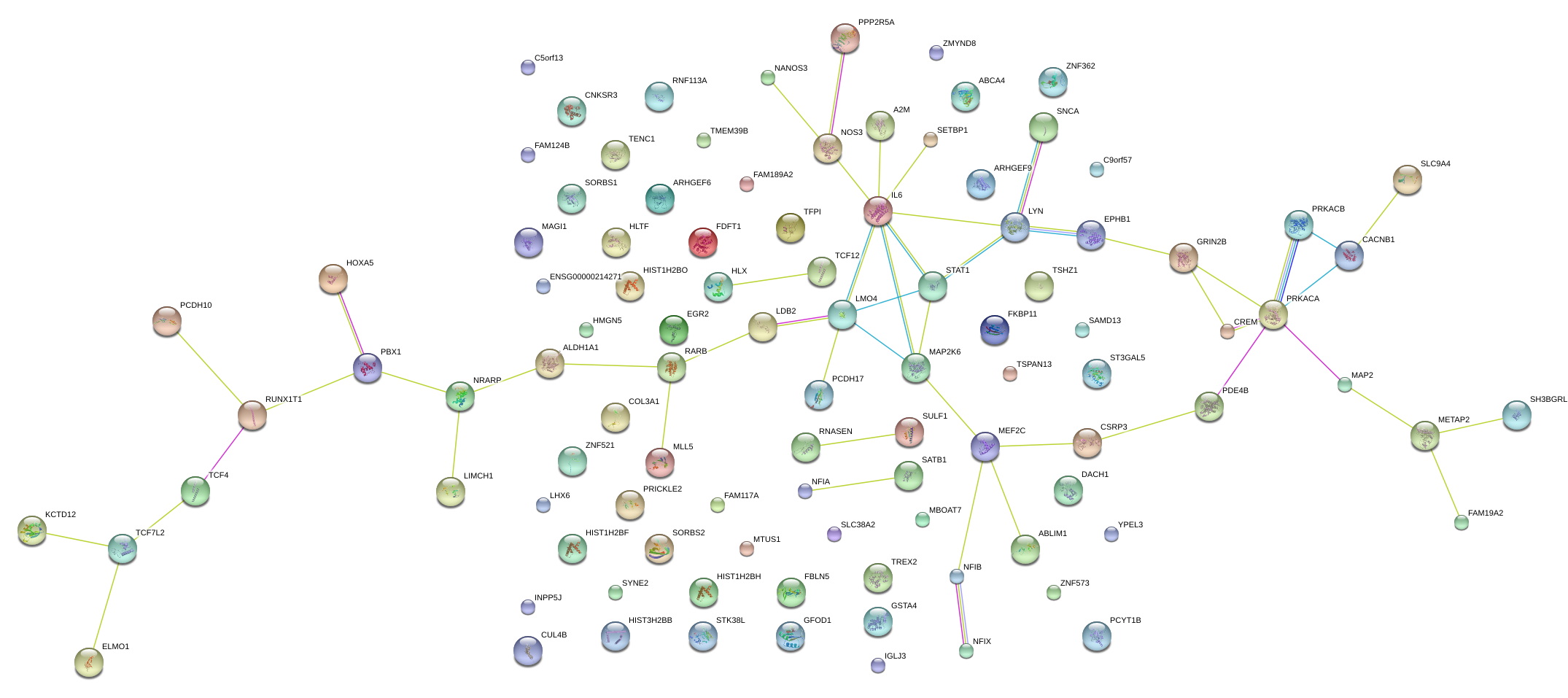
|
chr4_+_41700736
|
16.213
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr9_-_14086901
|
9.456
|
|
NFIB
|
nuclear factor I/B
|
|
chr1_+_61542928
|
9.387
|
NM_001145511
|
NFIA
|
nuclear factor I/A
|
|
chr4_-_186733372
|
6.326
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr3_-_64211111
|
6.007
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr1_+_61547388
|
5.959
|
NM_001145512
|
NFIA
|
nuclear factor I/A
|
|
chr4_+_41614933
|
5.104
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr4_+_41614911
|
5.016
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr1_+_164528586
|
4.962
|
NM_001204961
NM_001204963
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr4_+_41614725
|
4.934
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr18_-_53089722
|
4.898
|
NM_001243233
|
TCF4
|
transcription factor 4
|
|
chr1_+_84609941
|
4.825
|
NM_182948
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr8_-_93111915
|
4.693
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr17_+_67410825
|
4.459
|
NM_002758
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr5_-_88119604
|
4.140
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr1_+_61547625
|
3.667
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr13_-_77460432
|
3.646
|
NM_138444
|
KCTD12
|
potassium channel tetramerisation domain containing 12
|
|
chr10_-_116286661
|
3.521
|
NM_006720
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr3_-_18480203
|
3.328
|
NM_001131010
|
SATB1
|
SATB homeobox 1
|
|
chr1_+_228645807
|
3.142
|
NM_175055
|
HIST3H2BB
|
histone cluster 3, H2bb
|
|
chr2_-_225266710
|
2.953
|
NM_001122779
NM_024785
|
FAM124B
|
family with sequence similarity 124B
|
|
chr12_+_53442732
|
2.940
|
NM_015319
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr13_+_58205788
|
2.935
|
NM_001040429
|
PCDH17
|
protocadherin 17
|
|
chr18_-_22930826
|
2.817
|
|
|
|
|
chr18_+_72922725
|
2.808
|
NM_005786
|
TSHZ1
|
teashirt zinc finger homeobox 1
|
|
chr7_+_150690838
|
2.639
|
NM_001160109
NM_001160110
NM_001160111
|
NOS3
|
nitric oxide synthase 3 (endothelial cell)
|
|
chr5_-_111091947
|
2.635
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr1_+_164528914
|
2.576
|
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr3_+_25470067
|
2.534
|
|
RARB
|
retinoic acid receptor, beta
|
|
chr10_-_116392099
|
2.474
|
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr18_-_22932109
|
2.467
|
NM_015461
|
ZNF521
|
zinc finger protein 521
|
|
chr1_+_84767288
|
2.443
|
NM_001134663
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr18_+_42260774
|
2.431
|
NM_015559
|
SETBP1
|
SET binding protein 1
|
|
chr4_+_134070443
|
2.296
|
NM_020815
NM_032961
|
PCDH10
|
protocadherin 10
|
|
chr2_-_28113954
|
2.266
|
|
LOC100302650
|
uncharacterized LOC100302650
|
|
chr8_-_93107442
|
2.255
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr14_-_92413738
|
2.181
|
|
FBLN5
|
fibulin 5
|
|
chr7_-_27183225
|
2.150
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr18_+_8717368
|
2.127
|
NM_015210
|
CCDC165
|
coiled-coil domain containing 165
|
|
chr10_-_64578926
|
2.094
|
NM_001136178
|
EGR2
|
early growth response 2
|
|
chr14_-_92414004
|
2.069
|
NM_006329
|
FBLN5
|
fibulin 5
|
|
chr14_+_64682671
|
2.055
|
NM_182910
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2
|
|
chr9_+_71944240
|
2.025
|
NM_004816
|
FAM189A2
|
family with sequence similarity 189, member A2
|
|
chr5_-_88179278
|
2.014
|
NM_001193350
NM_002397
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr3_+_134514044
|
2.014
|
NM_004441
|
EPHB1
|
EPH receptor B1
|
|
chr9_-_140196702
|
1.976
|
NM_001004354
|
NRARP
|
NOTCH-regulated ankyrin repeat protein
|
|
chr3_-_148804122
|
1.975
|
NM_003071
NM_139048
|
HLTF
|
helicase-like transcription factor
|
|
chr10_-_64576074
|
1.962
|
|
EGR2
|
early growth response 2
|
|
chr10_-_64576123
|
1.947
|
NM_000399
NM_001136177
NM_001136179
|
EGR2
|
early growth response 2
|
|
chr4_-_90759446
|
1.874
|
NM_001146055
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor)
|
|
chr2_+_189839132
|
1.825
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr1_+_212475147
|
1.823
|
NM_001199756
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha
|
|
chr9_-_75567915
|
1.821
|
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|
|
chr12_+_27397077
|
1.797
|
NM_015000
|
STK38L
|
serine/threonine kinase 38 like
|
|
chr9_-_75568232
|
1.791
|
NM_000689
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|
|
chr10_-_97200895
|
1.785
|
NM_001034954
NM_001034955
NM_001034956
NM_001034957
NM_006434
|
SORBS1
|
sorbin and SH3 domain containing 1
|
|
chr1_+_33722158
|
1.784
|
NM_152493
|
ZNF362
|
zinc finger protein 362
|
|
chr1_+_221052845
|
1.762
|
|
HLX
|
H2.0-like homeobox
|
|
chr10_-_116286570
|
1.749
|
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr17_-_47841468
|
1.729
|
|
FAM117A
|
family with sequence similarity 117, member A
|
|
chr17_-_47841517
|
1.728
|
NM_030802
|
FAM117A
|
family with sequence similarity 117, member A
|
|
chr2_-_188419049
|
1.673
|
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr8_+_56792392
|
1.664
|
|
LYN
|
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog
|
|
chr22_+_31518923
|
1.661
|
NM_001002837
|
INPP5J
|
inositol polyphosphate-5-phosphatase J
|
|
chr13_-_72440143
|
1.630
|
|
DACH1
|
dachshund homolog 1 (Drosophila)
|
|
chr4_-_16900028
|
1.629
|
|
LDB2
|
LIM domain binding 2
|
|
chr2_-_188419185
|
1.589
|
NM_001032281
NM_006287
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr20_-_45927496
|
1.577
|
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chrX_-_135863104
|
1.571
|
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chrX_-_135863033
|
1.560
|
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chrX_-_24665225
|
1.554
|
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr13_-_72440557
|
1.544
|
|
DACH1
|
dachshund homolog 1 (Drosophila)
|
|
chr8_+_56792345
|
1.543
|
NM_001111097
NM_002350
|
LYN
|
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog
|
|
chr15_+_57511616
|
1.510
|
NM_207040
|
TCF12
|
transcription factor 12
|
|
chr9_-_74675498
|
1.482
|
NM_001128618
|
C9orf57
|
chromosome 9 open reading frame 57
|
|
chr1_+_66458389
|
1.475
|
NM_001037340
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr18_-_52988916
|
1.468
|
NM_001243234
NM_001243235
|
TCF4
|
transcription factor 4
|
|
chr7_+_104681378
|
1.467
|
|
MLL5
|
myeloid/lymphoid or mixed-lineage leukemia 5 (trithorax homolog, Drosophila)
|
|
chr10_+_35456443
|
1.466
|
NM_182771
NM_182772
|
CREM
|
cAMP responsive element modulator
|
|
chr1_+_15272414
|
1.452
|
NM_001017999
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr9_-_124990706
|
1.442
|
NM_001242333
|
LHX6
|
LIM homeobox 6
|
|
chr2_+_103089761
|
1.432
|
NM_001011552
|
SLC9A4
|
solute carrier family 9 (sodium/hydrogen exchanger), member 4
|
|
chr7_+_22766765
|
1.430
|
NM_000600
|
IL6
|
interleukin 6 (interferon, beta 2)
|
|
chrX_-_62974341
|
1.409
|
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chr5_-_111092918
|
1.387
|
NM_004772
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr6_-_13408368
|
1.386
|
NM_001242630
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1
|
|
chr1_+_32538497
|
1.384
|
NM_018056
|
TMEM39B
|
transmembrane protein 39B
|
|
chr16_-_30107492
|
1.338
|
NM_031477
NM_001145524
|
YPEL3
|
yippee-like 3 (Drosophila)
|
|
chr7_+_22767101
|
1.322
|
|
IL6
|
interleukin 6 (interferon, beta 2)
|
|
chr2_-_86094792
|
1.308
|
NM_001042437
|
ST3GAL5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5
|
|
chrX_+_80457597
|
1.307
|
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like
|
|
chr12_-_46766019
|
1.274
|
|
SLC38A2
|
solute carrier family 38, member 2
|
|
chr8_+_11665394
|
1.244
|
|
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1
|
|
chr5_-_88199868
|
1.236
|
NM_001131005
NM_001193347
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr3_-_66024508
|
1.230
|
NM_001033057
NM_004742
NM_015520
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1
|
|
chr1_+_32538521
|
1.229
|
|
TMEM39B
|
transmembrane protein 39B
|
|
chr1_-_94586678
|
1.223
|
NM_000350
|
ABCA4
|
ATP-binding cassette, sub-family A (ABC1), member 4
|
|
chrX_-_119694492
|
1.222
|
|
CUL4B
|
cullin 4B
|
|
chr1_+_221053000
|
1.218
|
|
HLX
|
H2.0-like homeobox
|
|
chr8_+_70405080
|
1.211
|
|
SULF1
|
sulfatase 1
|
|
chr18_-_53255734
|
1.205
|
NM_001083962
NM_001243230
NM_003199
|
TCF4
|
transcription factor 4
|
|
chrX_-_135863502
|
1.195
|
NM_004840
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr8_-_93107643
|
1.191
|
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr2_-_191872322
|
1.184
|
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa
|
|
chr19_-_54692132
|
1.184
|
|
MBOAT7
|
membrane bound O-acyltransferase domain containing 7
|
|
chr2_+_210444364
|
1.180
|
NM_002374
NM_031845
NM_031847
|
MAP2
|
microtubule-associated protein 2
|
|
chr12_-_14132920
|
1.179
|
NM_000834
|
GRIN2B
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2B
|
|
chr5_-_111312522
|
1.176
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr7_-_37024664
|
1.166
|
NM_001039459
|
ELMO1
|
engulfment and cell motility 1
|
|
chr7_+_16793320
|
1.158
|
NM_014399
|
TSPAN13
|
tetraspanin 13
|
|
chr1_+_87797371
|
1.156
|
|
LMO4
|
LIM domain only 4
|
|
chr5_-_31532164
|
1.152
|
NM_001100412
NM_013235
|
DROSHA
|
drosha, ribonuclease type III
|
|
chr8_-_17555158
|
1.146
|
NM_020749
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr22_+_23063107
|
1.140
|
|
IGLJ3
|
immunoglobulin lambda joining 3
|
|
chr8_+_11665959
|
1.137
|
|
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1
|
|
chr2_+_189839078
|
1.134
|
NM_000090
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr12_-_9268522
|
1.132
|
NM_000014
|
A2M
|
alpha-2-macroglobulin
|
|
chrX_-_80457440
|
1.129
|
NM_030763
|
HMGN5
|
high mobility group nucleosome binding domain 5
|
|
chr6_-_52860076
|
1.124
|
NM_001512
|
GSTA4
|
glutathione S-transferase alpha 4
|
|
chr18_-_53255361
|
1.123
|
|
TCF4
|
transcription factor 4
|
|
chr8_+_56792399
|
1.111
|
|
LYN
|
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog
|
|
chrX_+_80457559
|
1.110
|
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like
|
|
chr4_-_159094163
|
1.104
|
NM_001031700
NM_001128424
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr6_+_148663953
|
1.101
|
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr14_-_21271002
|
1.098
|
NM_002933
NM_198234
NM_198235
|
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic)
|
|
chr12_-_9268440
|
1.097
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr12_-_6772252
|
1.093
|
NM_001127582
NM_001127583
NM_001127584
NM_001127585
NM_001127586
NM_016162
|
ING4
|
inhibitor of growth family, member 4
|
|
chr2_+_173686314
|
1.092
|
NM_001100397
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4
|
|
chr11_-_27494232
|
1.079
|
NM_018490
|
LGR4
|
leucine-rich repeat containing G protein-coupled receptor 4
|
|
chr1_-_21377387
|
1.079
|
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3
|
|
chrX_+_80457302
|
1.075
|
NM_003022
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like
|
|
chr5_+_68800003
|
1.074
|
NM_001205255
|
OCLN
|
occludin
|
|
chr6_+_148663728
|
1.072
|
NM_015278
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr1_+_84768333
|
1.063
|
NM_001134664
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr6_+_143929316
|
1.061
|
NM_001100166
NM_014721
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr14_+_53019865
|
1.061
|
NM_001099652
|
GPR137C
|
G protein-coupled receptor 137C
|
|
chrX_+_70338403
|
1.060
|
NM_005120
|
MED12
|
mediator complex subunit 12
|
|
chr3_+_109128836
|
1.059
|
NM_001145553
|
FLJ25363
|
uncharacterized LOC401082
|
|
chr5_+_153570270
|
1.057
|
NM_198321
|
GALNT10
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10)
|
|
chr17_-_8063946
|
1.055
|
|
VAMP2
|
vesicle-associated membrane protein 2 (synaptobrevin 2)
|
|
chr10_+_102107074
|
1.054
|
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase)
|
|
chr10_+_102106958
|
1.042
|
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase)
|
|
chr5_-_111092865
|
1.041
|
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr15_+_96869156
|
1.035
|
NM_001145155
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr20_+_39657612
|
1.033
|
|
TOP1
|
topoisomerase (DNA) I
|
|
chr8_-_93075190
|
1.032
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr6_+_148664124
|
1.017
|
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr22_+_25591072
|
1.012
|
|
KIAA1671
|
KIAA1671
|
|
chr4_+_140586921
|
1.003
|
NM_001204366
NM_001204367
NM_001204368
NM_002413
|
MGST2
|
microsomal glutathione S-transferase 2
|
|
chr18_-_53257033
|
0.990
|
NM_001243227
NM_001243228
|
TCF4
|
transcription factor 4
|
|
chr10_-_105452878
|
0.985
|
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr11_-_2906960
|
0.972
|
NM_000076
NM_001122630
NM_001122631
|
CDKN1C
|
cyclin-dependent kinase inhibitor 1C (p57, Kip2)
|
|
chr9_-_124990949
|
0.970
|
NM_014368
NM_199160
|
LHX6
|
LIM homeobox 6
|
|
chr20_-_33735060
|
0.968
|
NM_001145025
NM_018217
|
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2
|
|
chr11_-_66115016
|
0.967
|
NM_006876
|
B3GNT1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1
|
|
chr4_-_159093523
|
0.963
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr17_-_26127395
|
0.954
|
|
NOS2
|
nitric oxide synthase 2, inducible
|
|
chr13_-_48987247
|
0.948
|
|
LPAR6
|
lysophosphatidic acid receptor 6
|
|
chrX_+_28605558
|
0.935
|
NM_014271
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1
|
|
chr10_+_102107256
|
0.930
|
|
|
|
|
chr20_-_45976626
|
0.929
|
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr2_+_33359607
|
0.927
|
NM_000627
NM_001166264
NM_001166265
NM_001166266
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr10_+_102107001
|
0.927
|
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase)
|
|
chr3_+_134514300
|
0.925
|
|
EPHB1
|
EPH receptor B1
|
|
chr3_-_52479042
|
0.923
|
NM_020163
|
SEMA3G
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3G
|
|
chr15_+_96875793
|
0.923
|
NM_001145156
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr12_-_9268513
|
0.920
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr3_-_112359251
|
0.920
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr1_-_228645517
|
0.916
|
NM_033445
|
HIST3H2A
|
histone cluster 3, H2a
|
|
chr9_+_140172273
|
0.916
|
NM_017723
|
C9orf167
|
chromosome 9 open reading frame 167
|
|
chr12_-_50616425
|
0.915
|
NM_001113547
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr18_+_77623667
|
0.910
|
NM_012283
|
KCNG2
|
potassium voltage-gated channel, subfamily G, member 2
|
|
chr17_+_38219040
|
0.885
|
NM_001190918
NM_003250
NM_199334
|
THRA
|
thyroid hormone receptor, alpha
|
|
chr20_+_306205
|
0.878
|
NM_006943
|
SOX12
|
SRY (sex determining region Y)-box 12
|
|
chr4_-_159093717
|
0.878
|
NM_016613
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr20_+_39657461
|
0.875
|
NM_003286
|
TOP1
|
topoisomerase (DNA) I
|
|
chr5_-_95158465
|
0.874
|
NM_001118890
NM_001243658
NM_001243659
NM_002064
|
GLRX
|
glutaredoxin (thioltransferase)
|
|
chr17_-_26127474
|
0.872
|
NM_000625
|
NOS2
|
nitric oxide synthase 2, inducible
|
|
chr3_-_112359660
|
0.868
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr3_-_112359443
|
0.866
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr3_+_50192847
|
0.864
|
NM_004186
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F
|
|
chr2_-_192015733
|
0.862
|
|
STAT4
|
signal transducer and activator of transcription 4
|
|
chr7_+_129007942
|
0.860
|
NM_001130723
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr8_+_70405027
|
0.858
|
NM_001128205
NM_001128206
|
SULF1
|
sulfatase 1
|
|
chr3_+_50192420
|
0.856
|
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F
|
|
chr20_-_33734894
|
0.848
|
|
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2
|
|
chr6_+_136172802
|
0.831
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr7_+_74188308
|
0.828
|
NM_000265
|
NCF1
NCF1B
|
neutrophil cytosolic factor 1
neutrophil cytosolic factor 1B pseudogene
|
|
chr3_+_49568370
|
0.828
|
|
DAG1
|
dystroglycan 1 (dystrophin-associated glycoprotein 1)
|
|
chr3_-_112359565
|
0.826
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr6_+_52935770
|
0.821
|
NM_012347
|
FBXO9
|
F-box protein 9
|
|
chr1_-_149783910
|
0.820
|
NM_001024599
NM_001161334
|
HIST2H2BF
|
histone cluster 2, H2bf
|
|
chr1_-_211752052
|
0.817
|
NM_021194
|
SLC30A1
|
solute carrier family 30 (zinc transporter), member 1
|
|
chrX_-_33357725
|
0.814
|
NM_000109
|
DMD
|
dystrophin
|
|
chr1_-_21377452
|
0.809
|
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3
|
|
chr4_-_141075232
|
0.806
|
NM_018717
|
MAML3
|
mastermind-like 3 (Drosophila)
|
|
chr11_+_5653484
|
0.805
|
NM_130390
|
TRIM34
|
tripartite motif containing 34
|
|
chr13_-_48987017
|
0.801
|
|
LPAR6
|
lysophosphatidic acid receptor 6
|
|
chrX_-_117251302
|
0.796
|
NM_001168301
|
KLHL13
|
kelch-like 13 (Drosophila)
|
|
chr2_+_109384601
|
0.792
|
|
RANBP2
|
RAN binding protein 2
|