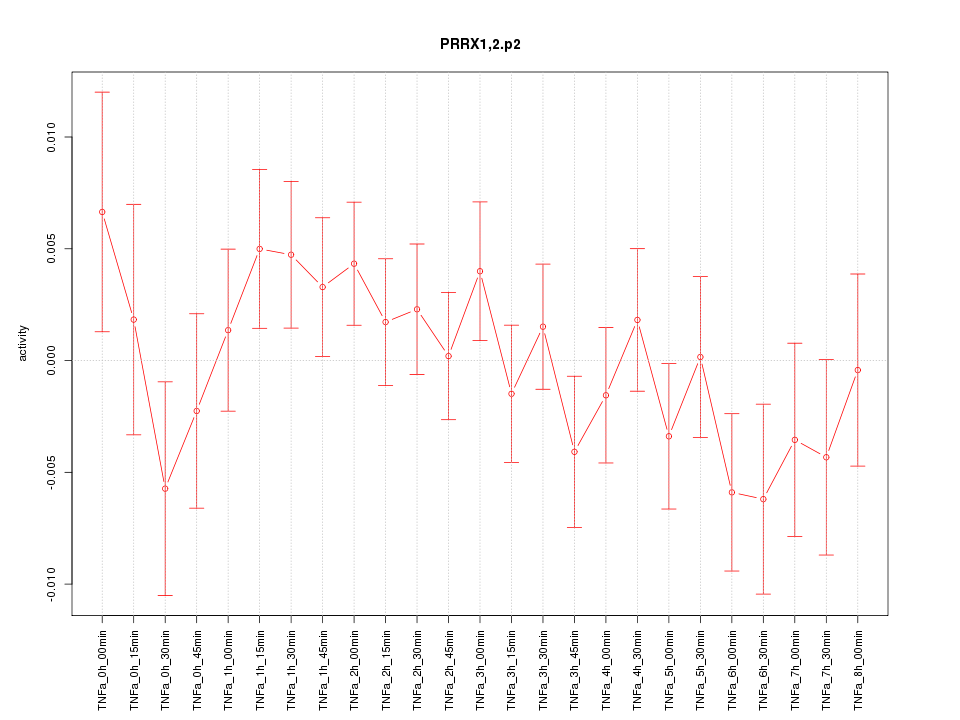
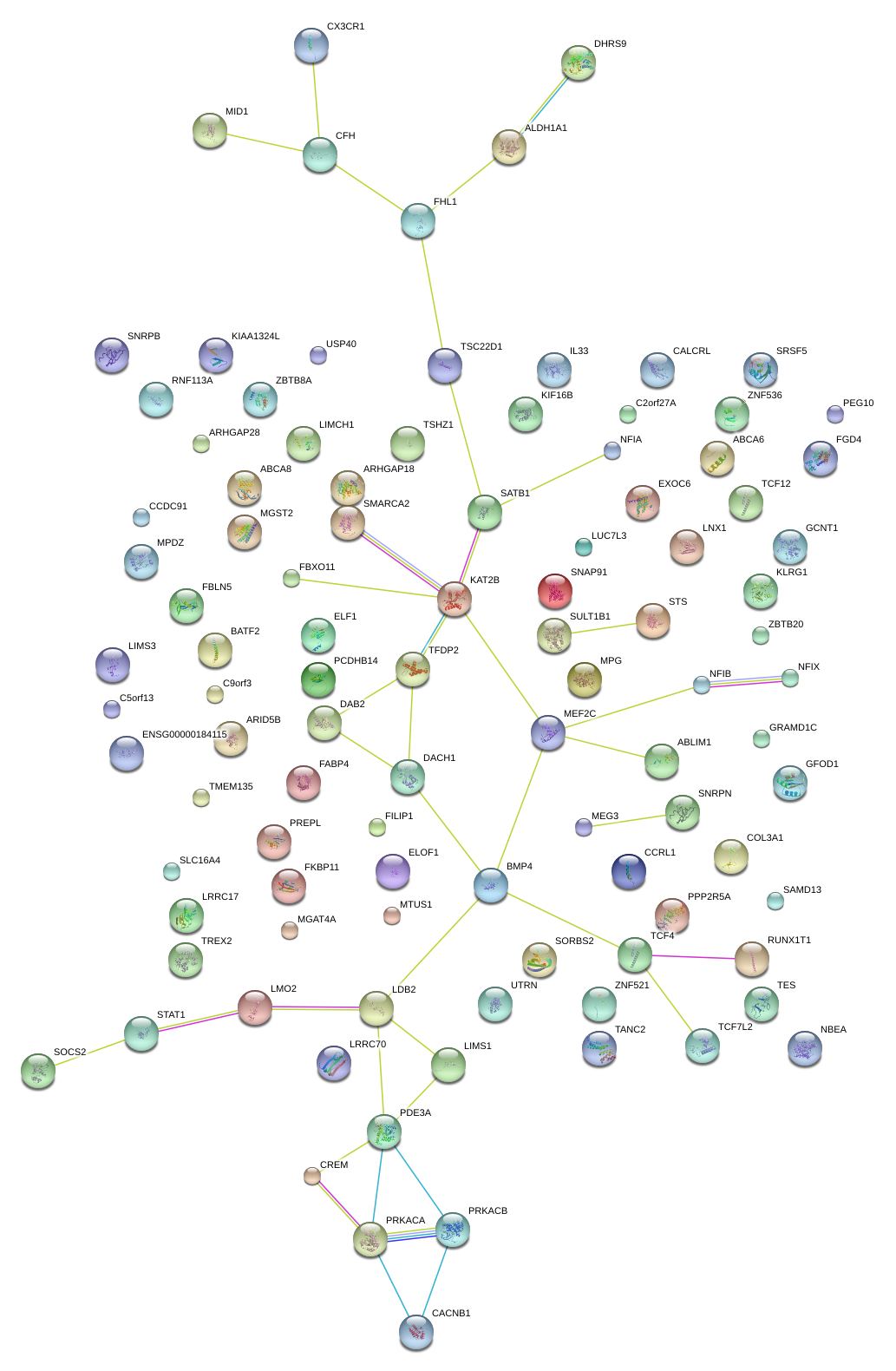
|
chr1_+_84630575
|
7.876
|
NM_001242860
NM_001242861
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr9_-_14086901
|
6.849
|
|
NFIB
|
nuclear factor I/B
|
|
chr5_-_88119604
|
5.740
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr15_+_25281105
|
4.114
|
|
SNRPN
|
small nuclear ribonucleoprotein polypeptide N
|
|
chr1_+_84647342
|
4.034
|
NM_001242862
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr1_+_84630014
|
3.889
|
NM_001242857
NM_001242858
NM_001242859
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr4_+_41614725
|
3.578
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr9_-_14398981
|
3.106
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chr11_+_87032428
|
3.034
|
|
TMEM135
|
transmembrane protein 135
|
|
chr3_-_18480203
|
2.951
|
NM_001131010
|
SATB1
|
SATB homeobox 1
|
|
chr8_-_17555158
|
2.942
|
NM_020749
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr5_-_88199868
|
2.757
|
NM_001131005
NM_001193347
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr5_-_88179278
|
2.649
|
NM_001193350
NM_002397
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr1_+_84630374
|
2.647
|
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr13_+_36050885
|
2.616
|
NM_001204197
|
NBEA
|
neurobeachin
|
|
chr4_+_41540187
|
2.522
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr17_-_67138013
|
2.518
|
NM_080284
|
ABCA6
|
ATP-binding cassette, sub-family A (ABC1), member 6
|
|
chr2_-_188312872
|
2.479
|
NM_005795
|
CALCRL
|
calcitonin receptor-like
|
|
chr18_-_53089722
|
2.470
|
NM_001243233
|
TCF4
|
transcription factor 4
|
|
chr7_+_115863004
|
2.409
|
NM_152829
|
TES
|
testis derived transcript (3 LIM domains)
|
|
chr6_-_130031278
|
2.368
|
NM_033515
|
ARHGAP18
|
Rho GTPase activating protein 18
|
|
chr4_+_41540320
|
2.355
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr13_-_72441312
|
2.348
|
NM_004392
NM_080759
NM_080760
|
DACH1
|
dachshund homolog 1 (Drosophila)
|
|
chr10_-_116392099
|
2.283
|
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr4_+_41614933
|
2.282
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr4_-_16900348
|
2.277
|
NM_001130834
NM_001290
|
LDB2
|
LIM domain binding 2
|
|
chr4_-_16900256
|
2.253
|
|
LDB2
|
LIM domain binding 2
|
|
chr4_-_16900028
|
2.249
|
|
LDB2
|
LIM domain binding 2
|
|
chr4_+_41614911
|
2.233
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr4_+_140587086
|
2.164
|
|
MGST2
|
microsomal glutathione S-transferase 2
|
|
chr4_+_41700736
|
2.131
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr10_+_94594469
|
2.057
|
NM_001013848
|
EXOC6
|
exocyst complex component 6
|
|
chr1_+_61547625
|
2.056
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr9_-_13250313
|
2.051
|
NM_003829
|
MPDZ
|
multiple PDZ domain protein
|
|
chr1_+_61547388
|
2.024
|
NM_001145512
|
NFIA
|
nuclear factor I/A
|
|
chr1_+_84768333
|
1.999
|
NM_001134664
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr8_-_82395401
|
1.927
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr17_+_48821106
|
1.895
|
|
LUC7L3
|
LUC7-like 3 (S. cerevisiae)
|
|
chr6_-_84419018
|
1.879
|
|
SNAP91
|
synaptosomal-associated protein, 91kDa homolog (mouse)
|
|
chr12_+_9142136
|
1.867
|
NM_005810
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1
|
|
chr7_+_102553343
|
1.859
|
NM_001031692
NM_005824
|
LRRC17
|
leucine rich repeat containing 17
|
|
chr10_-_116418056
|
1.824
|
NM_002313
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr8_-_17579729
|
1.813
|
NM_001001931
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr6_-_76203256
|
1.780
|
NM_015687
|
FILIP1
|
filamin A interacting protein 1
|
|
chr1_+_61869699
|
1.778
|
|
NFIA
|
nuclear factor I/A
|
|
chr6_-_84419108
|
1.760
|
NM_001242793
NM_001242794
NM_014841
|
SNAP91
|
synaptosomal-associated protein, 91kDa homolog (mouse)
|
|
chr9_+_6215785
|
1.725
|
NM_001199640
NM_001199641
NM_033439
|
IL33
|
interleukin 33
|
|
chr5_-_111312522
|
1.718
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr2_+_109204777
|
1.704
|
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr12_+_93963597
|
1.661
|
NM_003877
|
SOCS2
|
suppressor of cytokine signaling 2
|
|
chr10_+_35426709
|
1.658
|
NM_183011
NM_183012
|
CREM
|
cAMP responsive element modulator
|
|
chr14_-_92413738
|
1.644
|
|
FBLN5
|
fibulin 5
|
|
chr4_-_186733372
|
1.642
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr9_+_79115548
|
1.640
|
NM_001097636
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chr2_-_191872322
|
1.617
|
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa
|
|
chr11_-_33913835
|
1.610
|
NM_005574
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr6_+_144612872
|
1.593
|
NM_007124
|
UTRN
|
utrophin
|
|
chr12_+_20724797
|
1.588
|
NM_001244683
|
PDE3A
|
phosphodiesterase 3A, cGMP-inhibited
|
|
chr7_-_86595203
|
1.540
|
NM_152748
|
KIAA1324L
|
KIAA1324-like
|
|
chr3_+_132316080
|
1.538
|
NM_016557
|
CCRL1
|
chemokine (C-C motif) receptor-like 1
|
|
chr8_-_17580024
|
1.524
|
NM_001166393
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr17_+_61086897
|
1.514
|
NM_025185
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2
|
|
chr1_+_33004771
|
1.510
|
NM_001040441
|
ZBTB8A
|
zinc finger and BTB domain containing 8A
|
|
chr9_-_75568232
|
1.503
|
NM_000689
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|
|
chr2_-_99279935
|
1.485
|
NM_001160154
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chr15_+_57511616
|
1.472
|
NM_207040
|
TCF12
|
transcription factor 12
|
|
chr17_-_66951381
|
1.466
|
NM_007168
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8
|
|
chrX_+_135230736
|
1.431
|
NM_001159700
|
FHL1
|
four and a half LIM domains 1
|
|
chr14_-_54423448
|
1.422
|
NM_001202
NM_130850
|
BMP4
|
bone morphogenetic protein 4
|
|
chr2_-_44586854
|
1.383
|
NM_001042385
NM_001042386
|
PREPL
|
prolyl endopeptidase-like
|
|
chr19_+_30863318
|
1.317
|
NM_014717
|
ZNF536
|
zinc finger protein 536
|
|
chr1_+_212475147
|
1.288
|
NM_001199756
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha
|
|
chr8_-_93111915
|
1.278
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr13_-_45010937
|
1.268
|
|
TSC22D1
|
TSC22 domain family, member 1
|
|
chr7_+_94297448
|
1.266
|
|
PEG10
|
paternally expressed 10
|
|
chr18_+_72922725
|
1.252
|
NM_005786
|
TSHZ1
|
teashirt zinc finger homeobox 1
|
|
chr14_+_101295409
|
1.250
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr1_+_97260515
|
1.243
|
|
|
|
|
chrX_-_10535476
|
1.186
|
NM_001193278
NM_001193279
NM_001193280
NM_001193281
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr4_-_70626429
|
1.141
|
NM_014465
|
SULT1B1
|
sulfotransferase family, cytosolic, 1B, member 1
|
|
chr3_-_114478117
|
1.125
|
NM_001164344
NM_001164345
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr3_+_20081523
|
1.119
|
NM_003884
|
KAT2B
|
K(lysine) acetyltransferase 2B
|
|
chr1_-_110933683
|
1.118
|
NM_001201546
NM_001201547
NM_001201548
NM_001201549
NM_004696
|
SLC16A4
|
solute carrier family 16, member 4 (monocarboxylic acid transporter 5)
|
|
chr5_+_140602914
|
1.114
|
NM_018934
|
PCDHB14
|
protocadherin beta 14
|
|
chr2_+_189839132
|
1.112
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr18_-_22931970
|
1.109
|
|
ZNF521
|
zinc finger protein 521
|
|
chr18_+_6834431
|
1.108
|
NM_001010000
|
ARHGAP28
|
Rho GTPase activating protein 28
|
|
chr1_+_196621007
|
1.103
|
NM_000186
NM_001014975
|
CFH
|
complement factor H
|
|
chr12_+_32654976
|
1.096
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr9_-_75567915
|
1.093
|
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|
|
chr6_-_13408368
|
1.090
|
NM_001242630
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1
|
|
chr2_+_109237679
|
1.071
|
NM_001193484
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr2_-_234474197
|
1.071
|
NM_018218
|
USP40
|
ubiquitin specific peptidase 40
|
|
chr5_+_61874561
|
1.069
|
NM_181506
|
LRRC70
|
leucine rich repeat containing 70
|
|
chr2_+_132480063
|
1.055
|
NM_013310
|
C2orf27A
|
chromosome 2 open reading frame 27A
|
|
chr9_+_97521930
|
1.053
|
NM_001193329
|
C9orf3
|
chromosome 9 open reading frame 3
|
|
chr3_+_113616317
|
1.050
|
NM_001172105
|
GRAMD1C
|
GRAM domain containing 1C
|
|
chr18_-_22932109
|
1.050
|
NM_015461
|
ZNF521
|
zinc finger protein 521
|
|
chr12_+_28410132
|
1.025
|
NM_018318
|
CCDC91
|
coiled-coil domain containing 91
|
|
chrX_+_7137471
|
1.014
|
NM_000351
|
STS
|
steroid sulfatase (microsomal), isozyme S
|
|
chr10_+_63808968
|
1.011
|
NM_001244638
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr14_+_70236266
|
1.007
|
|
SRSF5
|
serine/arginine-rich splicing factor 5
|
|
chr2_-_48115846
|
0.998
|
NM_025133
|
FBXO11
|
F-box protein 11
|
|
chr3_-_141747434
|
0.982
|
NM_001178141
NM_001178142
NM_006286
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr9_+_2015218
|
0.979
|
NM_003070
NM_139045
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2
|
|
chr1_+_33004969
|
0.976
|
|
ZBTB8A
|
zinc finger and BTB domain containing 8A
|
|
chr10_+_102123420
|
0.968
|
|
|
|
|
chr2_+_169937616
|
0.951
|
NM_001142271
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr9_-_14314036
|
0.947
|
NM_001190737
NM_005596
|
NFIB
|
nuclear factor I/B
|
|
chr5_-_39394423
|
0.946
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr13_-_41556188
|
0.939
|
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr12_+_29376596
|
0.922
|
NM_018099
|
FAR2
|
fatty acyl CoA reductase 2
|
|
chr15_+_93443559
|
0.906
|
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chrX_+_28605558
|
0.906
|
NM_014271
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1
|
|
chr17_-_67057046
|
0.905
|
NM_080283
|
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9
|
|
chr17_-_46622274
|
0.904
|
NM_002145
|
HOXB2
|
homeobox B2
|
|
chr3_+_46448678
|
0.885
|
NM_003965
NM_001130910
|
CCRL2
|
chemokine (C-C motif) receptor-like 2
|
|
chrX_+_13671224
|
0.880
|
NM_152634
|
TCEANC
|
transcription elongation factor A (SII) N-terminal and central domain containing
|
|
chr3_+_136649316
|
0.877
|
NM_001190796
|
NCK1
|
NCK adaptor protein 1
|
|
chr11_+_92085261
|
0.874
|
NM_001008781
|
FAT3
|
FAT tumor suppressor homolog 3 (Drosophila)
|
|
chrX_-_135749436
|
0.872
|
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr1_-_110933635
|
0.868
|
|
SLC16A4
|
solute carrier family 16, member 4 (monocarboxylic acid transporter 5)
|
|
chr9_+_71944240
|
0.867
|
NM_004816
|
FAM189A2
|
family with sequence similarity 189, member A2
|
|
chr21_-_39870303
|
0.859
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr1_+_100316530
|
0.857
|
NM_000646
|
AGL
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase
|
|
chr5_-_168271852
|
0.854
|
|
|
|
|
chr7_+_77469446
|
0.852
|
NM_001127357
NM_001127359
NM_020432
|
PHTF2
|
putative homeodomain transcription factor 2
|
|
chr12_+_15699277
|
0.852
|
NM_030668
NM_030669
NM_030670
NM_030671
|
PTPRO
|
protein tyrosine phosphatase, receptor type, O
|
|
chrX_-_117107700
|
0.833
|
NM_033495
NM_001168300
|
KLHL13
|
kelch-like 13 (Drosophila)
|
|
chr2_-_165424993
|
0.831
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr9_+_82187469
|
0.831
|
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chr12_-_29937691
|
0.830
|
NM_175861
|
TMTC1
|
transmembrane and tetratricopeptide repeat containing 1
|
|
chr20_-_45976626
|
0.829
|
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr4_-_138453564
|
0.824
|
|
PCDH18
|
protocadherin 18
|
|
chr16_+_53133063
|
0.819
|
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr7_-_108142934
|
0.817
|
|
PNPLA8
|
patatin-like phospholipase domain containing 8
|
|
chr2_+_128928040
|
0.817
|
|
UGGT1
|
UDP-glucose glycoprotein glucosyltransferase 1
|
|
chr2_+_109204904
|
0.812
|
NM_001193488
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr5_-_94417185
|
0.812
|
|
MCTP1
|
multiple C2 domains, transmembrane 1
|
|
chr2_+_179184970
|
0.811
|
NM_145739
|
OSBPL6
|
oxysterol binding protein-like 6
|
|
chr2_-_183387063
|
0.803
|
NM_005019
NM_001003683
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent
|
|
chr4_-_159094163
|
0.794
|
NM_001031700
NM_001128424
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr20_+_42574535
|
0.790
|
NM_001098798
|
TOX2
|
TOX high mobility group box family member 2
|
|
chr7_-_27183225
|
0.790
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr12_-_50616425
|
0.788
|
NM_001113547
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr4_-_152147520
|
0.786
|
NM_001009555
NM_001128923
|
SH3D19
|
SH3 domain containing 19
|
|
chr5_+_175108463
|
0.778
|
NM_022304
|
HRH2
|
histamine receptor H2
|
|
chr3_-_64211111
|
0.770
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chrX_-_18690187
|
0.768
|
NM_000330
|
RS1
|
retinoschisin 1
|
|
chr2_+_210444364
|
0.767
|
NM_002374
NM_031845
NM_031847
|
MAP2
|
microtubule-associated protein 2
|
|
chr12_-_7848324
|
0.766
|
NM_020634
|
GDF3
|
growth differentiation factor 3
|
|
chr12_-_10021932
|
0.764
|
|
CLEC2B
|
C-type lectin domain family 2, member B
|
|
chr2_+_189839078
|
0.764
|
NM_000090
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr10_+_35464514
|
0.761
|
NM_182769
NM_182770
|
CREM
|
cAMP responsive element modulator
|
|
chr2_+_173686314
|
0.759
|
NM_001100397
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4
|
|
chr1_+_145524989
|
0.756
|
NM_003637
|
ITGA10
|
integrin, alpha 10
|
|
chr4_-_186578122
|
0.754
|
NM_001145674
NM_001145675
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr2_-_188419049
|
0.750
|
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr9_+_12774988
|
0.749
|
NM_203403
|
C9orf150
|
chromosome 9 open reading frame 150
|
|
chr20_+_33759773
|
0.748
|
NM_006404
|
PROCR
|
protein C receptor, endothelial
|
|
chr5_+_65372744
|
0.748
|
|
ERBB2IP
|
erbb2 interacting protein
|
|
chr3_-_112358510
|
0.746
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr10_-_74283693
|
0.744
|
NM_001195519
|
MICU1
|
mitochondrial calcium uptake 1
|
|
chr7_+_129015483
|
0.743
|
NM_001130722
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr6_-_133119625
|
0.739
|
NM_052831
|
C6orf192
|
chromosome 6 open reading frame 192
|
|
chr12_+_101988746
|
0.737
|
NM_002465
NM_206819
NM_206820
NM_206821
|
MYBPC1
|
myosin binding protein C, slow type
|
|
chr18_+_33234532
|
0.737
|
NM_020474
|
GALNT1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 (GalNAc-T1)
|
|
chr4_-_152149042
|
0.737
|
NM_001243349
|
SH3D19
|
SH3 domain containing 19
|
|
chr6_+_153019029
|
0.735
|
NM_025107
|
MYCT1
|
myc target 1
|
|
chr3_-_112358758
|
0.734
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr3_-_69037498
|
0.732
|
|
C3orf64
|
chromosome 3 open reading frame 64
|
|
chr18_-_21891469
|
0.728
|
NM_001242508
|
OSBPL1A
|
oxysterol binding protein-like 1A
|
|
chr5_+_115898218
|
0.725
|
|
|
|
|
chr6_+_155538096
|
0.723
|
NM_001010927
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2
|
|
chr3_+_25470067
|
0.719
|
|
RARB
|
retinoic acid receptor, beta
|
|
chr3_+_159557745
|
0.718
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr6_-_75970566
|
0.717
|
|
TMEM30A
|
transmembrane protein 30A
|
|
chr6_+_136172802
|
0.714
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr5_+_68710938
|
0.712
|
NM_001038603
NM_001244734
|
MARVELD2
|
MARVEL domain containing 2
|
|
chr8_-_93075190
|
0.708
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr12_+_74931550
|
0.708
|
NM_001136262
|
ATXN7L3B
|
ataxin 7-like 3B
|
|
chr5_+_95066628
|
0.707
|
NM_014899
|
RHOBTB3
|
Rho-related BTB domain containing 3
|
|
chr21_-_40033617
|
0.707
|
NM_001243432
NM_001136154
NM_004449
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr20_+_42544781
|
0.704
|
NM_001098796
NM_032883
|
TOX2
|
TOX high mobility group box family member 2
|
|
chr11_-_31531109
|
0.704
|
NM_144981
|
IMMP1L
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae)
|
|
chr2_+_177016112
|
0.700
|
NM_014621
|
HOXD4
|
homeobox D4
|
|
chr17_+_39382878
|
0.692
|
NM_031961
|
KRTAP9-2
|
keratin associated protein 9-2
|
|
chr6_-_32157511
|
0.691
|
NM_002586
|
PBX2
|
pre-B-cell leukemia homeobox 2
|
|
chr17_-_46688279
|
0.691
|
|
HOXB7
|
homeobox B7
|
|
chr3_-_15839674
|
0.688
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr5_+_140624916
|
0.688
|
|
PCDHB15
|
protocadherin beta 15
|
|
chr11_+_60552820
|
0.678
|
NM_206893
|
MS4A10
|
membrane-spanning 4-domains, subfamily A, member 10
|
|
chr12_-_10022457
|
0.677
|
NM_005127
|
CLEC2B
|
C-type lectin domain family 2, member B
|
|
chr3_+_141043054
|
0.676
|
NM_001080412
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr4_-_70725792
|
0.676
|
NM_005420
|
SULT1E1
|
sulfotransferase family 1E, estrogen-preferring, member 1
|
|
chr8_-_17752847
|
0.675
|
NM_147203
NM_201553
NM_004467
NM_201552
|
FGL1
|
fibrinogen-like 1
|
|
chr5_-_111091947
|
0.675
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr10_-_65028825
|
0.674
|
NM_004241
|
JMJD1C
|
jumonji domain containing 1C
|
|
chr11_-_102745666
|
0.672
|
NM_002426
|
MMP12
|
matrix metallopeptidase 12 (macrophage elastase)
|
|
chr9_-_77643307
|
0.668
|
NM_152420
|
C9orf41
|
chromosome 9 open reading frame 41
|