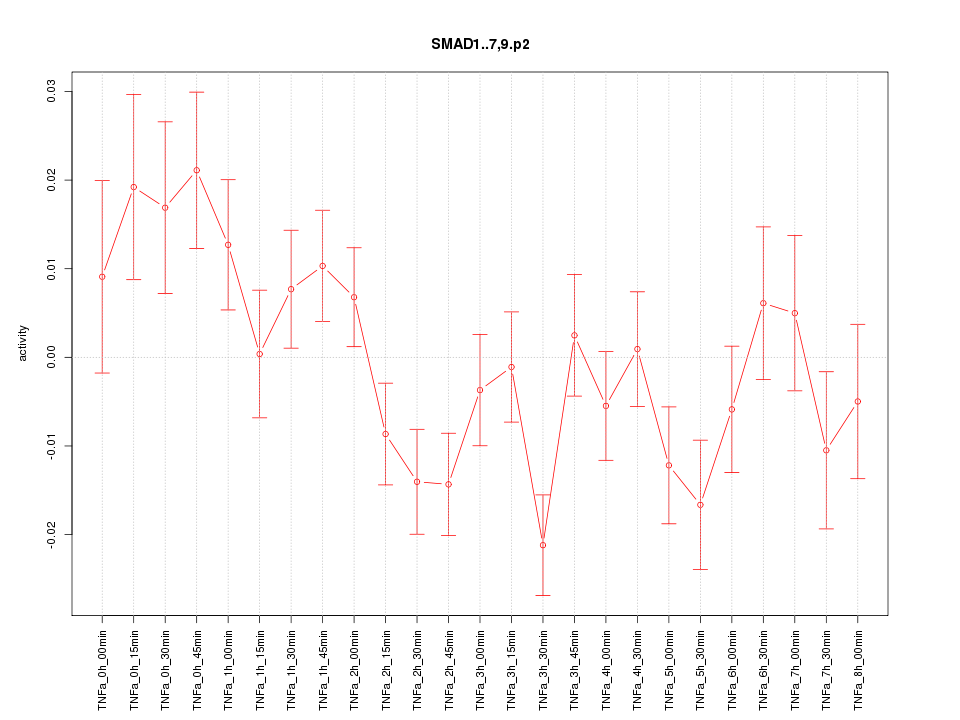
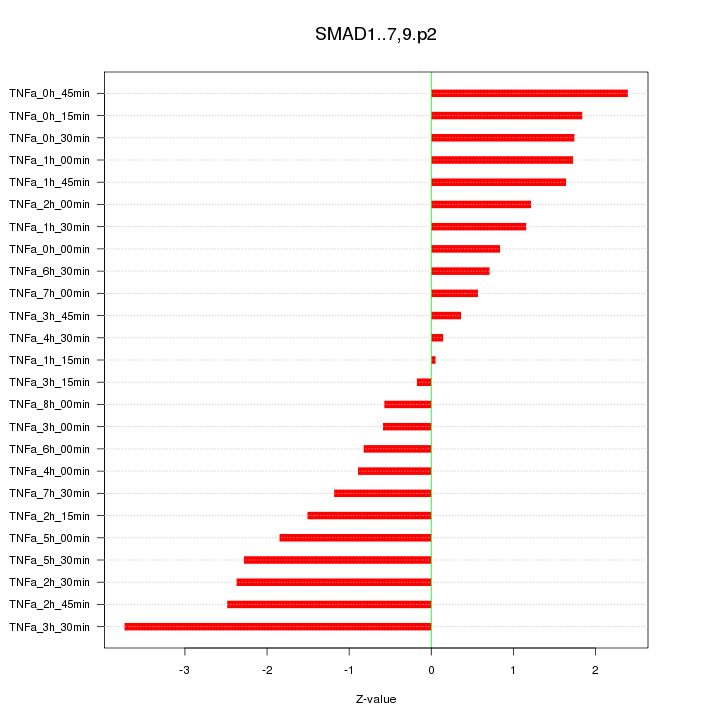
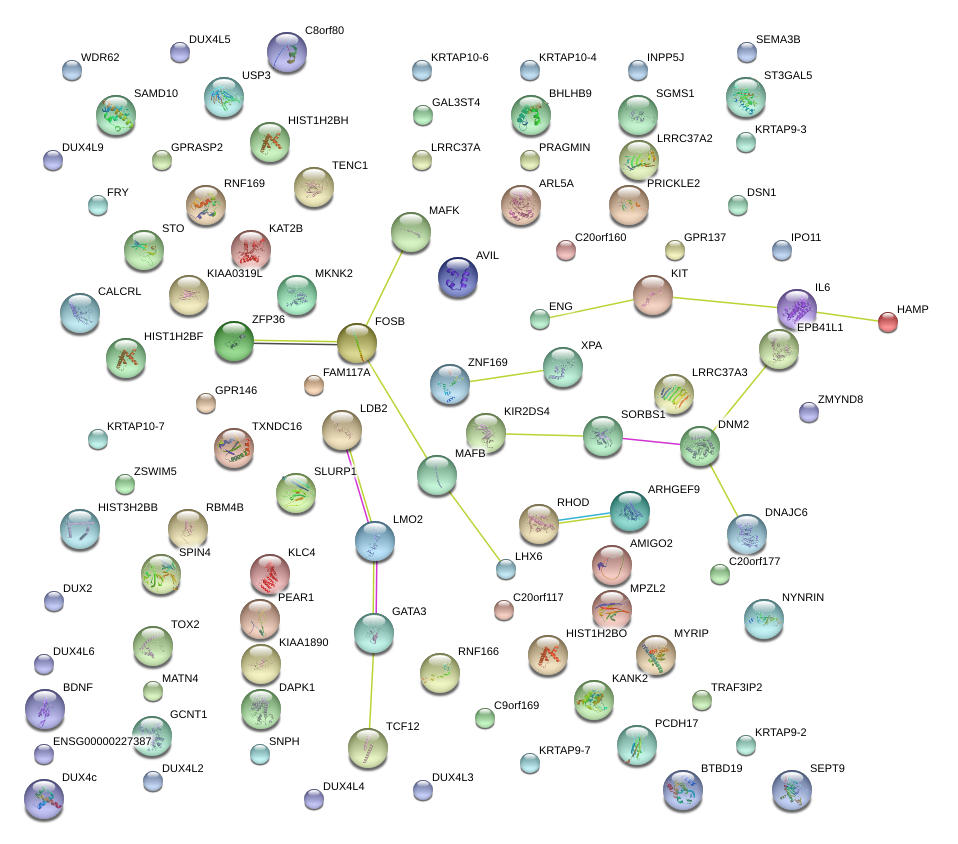
|
chr21_+_46020496
|
6.289
|
NM_198689
|
KRTAP10-7
|
keratin associated protein 10-7
|
|
chr12_+_53443834
|
4.644
|
NM_170754
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr13_+_32605436
|
3.558
|
NM_023037
|
FRY
|
furry homolog (Drosophila)
|
|
chr12_-_47473424
|
3.443
|
NM_001143668
NM_181847
|
AMIGO2
|
adhesion molecule with Ig-like domain 2
|
|
chrX_-_62570968
|
3.202
|
NM_001012968
|
SPIN4
|
spindlin family, member 4
|
|
chr21_-_46012383
|
3.140
|
NM_198688
|
KRTAP10-6
|
keratin associated protein 10-6
|
|
chr7_+_1097140
|
3.052
|
NM_138445
|
GPR146
|
G protein-coupled receptor 146
|
|
chr20_+_58515408
|
2.877
|
NM_001190827
NM_022106
|
C20orf177
|
chromosome 20 open reading frame 177
|
|
chr20_+_30598244
|
2.847
|
NM_080625
|
C20orf160
|
chromosome 20 open reading frame 160
|
|
chr8_-_8239188
|
2.828
|
NM_001080826
|
SGK223
|
homolog of rat pragma of Rnd2
|
|
chr22_+_31518923
|
2.622
|
NM_001002837
|
INPP5J
|
inositol polyphosphate-5-phosphatase J
|
|
chr9_-_100459556
|
2.616
|
|
XPA
|
xeroderma pigmentosum, complementation group A
|
|
chrX_+_101967290
|
2.570
|
|
GPRASP2
|
G protein-coupled receptor associated sorting protein 2
|
|
chr4_-_16900028
|
2.568
|
|
LDB2
|
LIM domain binding 2
|
|
chr21_+_45993605
|
2.489
|
NM_198687
|
KRTAP10-4
|
keratin associated protein 10-4
|
|
chr9_-_100459623
|
2.488
|
NM_000380
|
XPA
|
xeroderma pigmentosum, complementation group A
|
|
chr3_-_64211111
|
2.372
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr20_+_42543417
|
2.342
|
NM_001098797
|
TOX2
|
TOX high mobility group box family member 2
|
|
chr11_+_66824308
|
2.298
|
|
RHOD
|
ras homolog gene family, member D
|
|
chr2_-_188312872
|
2.293
|
NM_005795
|
CALCRL
|
calcitonin receptor-like
|
|
chr10_+_8096772
|
2.226
|
|
GATA3
|
GATA binding protein 3
|
|
chr4_-_16900348
|
2.193
|
NM_001130834
NM_001290
|
LDB2
|
LIM domain binding 2
|
|
chr20_-_45985411
|
2.182
|
NM_012408
NM_183047
NM_183048
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr16_-_88772807
|
2.166
|
NM_178841
|
RNF166
|
ring finger protein 166
|
|
chr10_+_8096633
|
2.112
|
NM_001002295
NM_002051
|
GATA3
|
GATA binding protein 3
|
|
chr11_-_33913835
|
2.104
|
NM_005574
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr20_+_1246959
|
2.098
|
NM_014723
|
SNPH
|
syntaphilin
|
|
chr20_-_45976626
|
2.085
|
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr16_-_88772732
|
2.067
|
NM_001171815
|
RNF166
|
ring finger protein 166
|
|
chr17_+_39382878
|
2.037
|
NM_031961
|
KRTAP9-2
|
keratin associated protein 9-2
|
|
chr12_+_53440809
|
1.951
|
NM_198316
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr11_-_33891485
|
1.938
|
NM_001142315
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr19_+_39897457
|
1.889
|
NM_003407
|
ZFP36
|
zinc finger protein 36, C3H type, homolog (mouse)
|
|
chr9_-_130617009
|
1.865
|
NM_000118
NM_001114753
|
ENG
|
endoglin
|
|
chr13_+_58205943
|
1.780
|
|
PCDH17
|
protocadherin 17
|
|
chr1_+_15272414
|
1.773
|
NM_001017999
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chrX_+_102000906
|
1.767
|
NM_001142529
NM_001142530
|
BHLHB9
|
basic helix-loop-helix domain containing, class B, 9
|
|
chr1_+_65720091
|
1.733
|
|
LOC645195
DNAJC6
|
uncharacterized LOC645195
DnaJ (Hsp40) homolog, subfamily C, member 6
|
|
chrX_-_62975005
|
1.719
|
NM_001173480
NM_015185
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chr20_-_39317875
|
1.700
|
NM_005461
|
MAFB
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian)
|
|
chr2_-_152684963
|
1.688
|
NM_001037174
NM_012097
NM_177985
|
ARL5A
|
ADP-ribosylation factor-like 5A
|
|
chr10_+_8097350
|
1.677
|
|
GATA3
|
GATA binding protein 3
|
|
chr17_-_5138049
|
1.649
|
NM_207103
|
SCIMP
|
SLP adaptor and CSK interacting membrane protein
|
|
chr1_+_22351984
|
1.648
|
|
LINC00339
|
long intergenic non-protein coding RNA 339
|
|
chr13_+_58205788
|
1.613
|
NM_001040429
|
PCDH17
|
protocadherin 17
|
|
chr20_-_35488275
|
1.613
|
NM_199181
|
KIAA0889
|
KIAA0889
|
|
chr3_+_20081853
|
1.595
|
|
KAT2B
|
K(lysine) acetyltransferase 2B
|
|
chr2_-_152684689
|
1.591
|
|
ARL5A
|
ADP-ribosylation factor-like 5A
|
|
chr19_-_2038937
|
1.563
|
|
MKNK2
|
MAP kinase interacting serine/threonine kinase 2
|
|
chr3_+_39851028
|
1.554
|
NM_015460
|
MYRIP
|
myosin VIIA and Rab interacting protein
|
|
chr11_+_74459900
|
1.552
|
NM_001098638
|
RNF169
|
ring finger protein 169
|
|
chr11_-_33891361
|
1.538
|
NM_001142316
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr19_-_11308184
|
1.504
|
NM_001136191
|
KANK2
|
KN motif and ankyrin repeat domains 2
|
|
chr20_+_34700257
|
1.489
|
NM_177996
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr1_+_228645807
|
1.476
|
NM_175055
|
HIST3H2BB
|
histone cluster 3, H2bb
|
|
chr12_-_58209840
|
1.475
|
NM_006576
|
AVIL
|
advillin
|
|
chr1_+_45274153
|
1.465
|
NM_001136537
|
BTBD19
|
BTB (POZ) domain containing 19
|
|
chr4_+_55523906
|
1.453
|
NM_000222
NM_001093772
|
KIT
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog
|
|
chr19_+_45971249
|
1.443
|
NM_001114171
NM_006732
|
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B
|
|
chr10_-_52383611
|
1.424
|
NM_147156
|
SGMS1
|
sphingomyelin synthase 1
|
|
chr11_-_33890931
|
1.401
|
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr9_-_124984018
|
1.386
|
NM_001242335
|
LHX6
|
LIM homeobox 6
|
|
chr3_+_50304989
|
1.372
|
NM_001005914
NM_004636
|
SEMA3B
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B
|
|
chr11_-_33891258
|
1.368
|
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr14_+_24867926
|
1.332
|
NM_025081
|
NYNRIN
|
NYN domain and retroviral integrase containing
|
|
chr19_+_35773409
|
1.294
|
NM_021175
|
HAMP
|
hepcidin antimicrobial peptide
|
|
chr17_-_62914902
|
1.258
|
NM_199340
|
LRRC37A3
|
leucine rich repeat containing 37, member A3
|
|
chr9_-_130616957
|
1.250
|
|
ENG
|
endoglin
|
|
chr7_+_22766765
|
1.249
|
NM_000600
|
IL6
|
interleukin 6 (interferon, beta 2)
|
|
chrX_+_101967103
|
1.239
|
NM_001004051
NM_001184874
NM_001184875
NM_001184876
NM_138437
|
GPRASP2
|
G protein-coupled receptor associated sorting protein 2
|
|
chr1_-_45672225
|
1.239
|
NM_020883
|
ZSWIM5
|
zinc finger, SWIM-type containing 5
|
|
chr15_+_63796793
|
1.227
|
NM_006537
|
USP3
|
ubiquitin specific peptidase 3
|
|
chr9_+_79056581
|
1.226
|
NM_001097634
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chrX_-_62974942
|
1.224
|
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chr20_-_35402142
|
1.219
|
NM_001145316
NM_024918
NM_001145315
NM_001145317
NM_001145318
|
DSN1
|
DSN1, MIND kinetochore complex component, homolog (S. cerevisiae)
|
|
chr1_-_36022938
|
1.215
|
|
KIAA0319L
|
KIAA0319-like
|
|
chr11_-_66445318
|
1.210
|
|
RBM4B
|
RNA binding motif protein 4B
|
|
chr11_-_66445218
|
1.209
|
NM_031492
|
RBM4B
|
RNA binding motif protein 4B
|
|
chr7_-_99766238
|
1.197
|
NM_024637
|
GAL3ST4
|
galactose-3-O-sulfotransferase 4
|
|
chr15_+_57210852
|
1.196
|
|
TCF12
|
transcription factor 12
|
|
chr10_+_135480367
|
1.196
|
NM_033178
NM_012147
NM_001127387
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L7
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 7
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr17_-_47841517
|
1.186
|
NM_030802
|
FAM117A
|
family with sequence similarity 117, member A
|
|
chr20_-_43936919
|
1.172
|
NM_003833
NM_030590
NM_030592
|
MATN4
|
matrilin 4
|
|
chr9_-_130616760
|
1.168
|
|
ENG
|
endoglin
|
|
chr11_+_66824285
|
1.166
|
NM_014578
|
RHOD
|
ras homolog gene family, member D
|
|
chr15_+_57210889
|
1.166
|
|
TCF12
|
transcription factor 12
|
|
chr6_+_43027628
|
1.149
|
NM_201523
|
KLC4
|
kinesin light chain 4
|
|
chr4_+_191001978
|
1.147
|
NM_001177376
|
DUX4L4
|
double homeobox 4 like 4
|
|
chr4_+_191008565
|
1.135
|
NM_012147
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX2
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 2
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr4_+_190995385
|
1.125
|
NM_012147
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX2
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 2
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr4_+_190998678
|
1.123
|
NM_012147
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX2
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 2
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr4_+_190988866
|
1.122
|
NM_012147
|
DUX2
|
double homeobox 2
|
|
chr10_+_135493659
|
1.121
|
NM_012147
|
DUX2
|
double homeobox 2
|
|
chr10_+_135490278
|
1.111
|
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr3_+_20081523
|
1.109
|
NM_003884
|
KAT2B
|
K(lysine) acetyltransferase 2B
|
|
chr9_+_90112754
|
1.107
|
NM_004938
|
DAPK1
|
death-associated protein kinase 1
|
|
chr4_+_191011859
|
1.106
|
NM_033178
NM_012147
NM_001177376
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L4
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr4_+_191005266
|
1.106
|
NM_033178
NM_012147
NM_001177376
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L4
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr9_+_140119643
|
1.094
|
NM_199001
|
C9orf169
|
chromosome 9 open reading frame 169
|
|
chr11_-_118135197
|
1.090
|
NM_005797
NM_144765
|
MPZL2
|
myelin protein zero-like 2
|
|
chr8_-_27941387
|
1.084
|
NM_001010906
|
C8orf80
|
chromosome 8 open reading frame 80
|
|
chr17_-_62915580
|
1.077
|
|
LRRC37A3
|
leucine rich repeat containing 37, member A3
|
|
chr2_-_86094792
|
1.074
|
NM_001042437
|
ST3GAL5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5
|
|
chr8_-_143823823
|
1.072
|
NM_020427
|
SLURP1
|
secreted LY6/PLAUR domain containing 1
|
|
chrX_+_101975641
|
1.066
|
NM_001142524
NM_001142525
NM_001142526
NM_001142527
NM_001142528
NM_030639
|
BHLHB9
|
basic helix-loop-helix domain containing, class B, 9
|
|
chr11_-_27741192
|
1.065
|
NM_001143807
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr10_-_97200895
|
1.062
|
NM_001034954
NM_001034955
NM_001034956
NM_001034957
NM_006434
|
SORBS1
|
sorbin and SH3 domain containing 1
|
|
chr11_-_62472901
|
1.047
|
|
|
|
|
chr20_-_62610990
|
1.047
|
NM_080621
|
SAMD10
|
sterile alpha motif domain containing 10
|
|
chr5_+_61714710
|
1.045
|
NM_001134779
|
IPO11
|
importin 11
|
|
chr11_-_118134956
|
1.032
|
|
MPZL2
|
myelin protein zero-like 2
|
|
chr6_+_43027368
|
1.028
|
NM_138343
NM_201521
|
KLC4
|
kinesin light chain 4
|
|
chr9_-_35111534
|
1.026
|
|
FAM214B
|
family with sequence similarity 214, member B
|
|
chr11_-_118134986
|
1.020
|
|
MPZL2
|
myelin protein zero-like 2
|
|
chr7_+_1581072
|
1.017
|
|
MAFK
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian)
|
|
chr5_+_176560056
|
1.006
|
NM_172349
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr19_-_11308457
|
1.003
|
|
KANK2
|
KN motif and ankyrin repeat domains 2
|
|
chr6_-_111927059
|
1.000
|
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chr1_+_156863522
|
0.992
|
NM_001080471
|
PEAR1
|
platelet endothelial aggregation receptor 1
|
|
chr2_-_110874142
|
0.987
|
NM_005434
|
MALL
|
mal, T-cell differentiation protein-like
|
|
chrX_-_62974987
|
0.983
|
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chr20_-_36152952
|
0.983
|
NM_001167823
|
BLCAP
|
bladder cancer associated protein
|
|
chr17_-_47841468
|
0.973
|
|
FAM117A
|
family with sequence similarity 117, member A
|
|
chr10_-_116392099
|
0.965
|
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr6_-_84419108
|
0.963
|
NM_001242793
NM_001242794
NM_014841
|
SNAP91
|
synaptosomal-associated protein, 91kDa homolog (mouse)
|
|
chr3_+_32148000
|
0.958
|
NM_015141
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like
|
|
chr19_-_41933255
|
0.953
|
|
|
|
|
chr11_+_64107687
|
0.947
|
NM_032251
|
CCDC88B
|
coiled-coil domain containing 88B
|
|
chr5_-_90679115
|
0.942
|
NM_020801
|
ARRDC3
|
arrestin domain containing 3
|
|
chr1_-_59012379
|
0.941
|
NM_145243
|
OMA1
|
OMA1 zinc metallopeptidase homolog (S. cerevisiae)
|
|
chr1_+_212459169
|
0.940
|
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha
|
|
chr12_+_52695648
|
0.930
|
NM_002284
|
KRT86
|
keratin 86
|
|
chr9_+_130374466
|
0.930
|
NM_001032221
NM_003165
|
STXBP1
|
syntaxin binding protein 1
|
|
chr15_+_57210823
|
0.922
|
NM_003205
NM_207036
NM_207037
NM_207038
|
TCF12
|
transcription factor 12
|
|
chr7_-_99766191
|
0.921
|
|
GAL3ST4
|
galactose-3-O-sulfotransferase 4
|
|
chr3_+_44626453
|
0.920
|
NM_173658
|
ZNF660
|
zinc finger protein 660
|
|
chr6_-_84419018
|
0.919
|
|
SNAP91
|
synaptosomal-associated protein, 91kDa homolog (mouse)
|
|
chr3_+_51976274
|
0.917
|
NM_001003931
NM_005485
|
PARP3
|
poly (ADP-ribose) polymerase family, member 3
|
|
chr4_+_30721865
|
0.913
|
NM_032456
NM_001173523
NM_002589
NM_032457
|
PCDH7
|
protocadherin 7
|
|
chr1_+_212459262
|
0.910
|
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha
|
|
chr3_+_32148136
|
0.906
|
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like
|
|
chr14_+_45605143
|
0.891
|
|
FANCM
|
Fanconi anemia, complementation group M
|
|
chr6_+_18387580
|
0.890
|
NM_182757
|
RNF144B
|
ring finger protein 144B
|
|
chr15_+_66995414
|
0.882
|
|
SMAD6
|
SMAD family member 6
|
|
chr9_-_130616592
|
0.882
|
|
ENG
|
endoglin
|
|
chr4_+_159131400
|
0.877
|
NM_018342
|
TMEM144
|
transmembrane protein 144
|
|
chr12_-_104443698
|
0.874
|
|
GLT8D2
|
glycosyltransferase 8 domain containing 2
|
|
chr13_+_33160537
|
0.867
|
NM_015032
|
PDS5B
|
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae)
|
|
chr22_+_29702950
|
0.866
|
NM_006478
NM_152236
NM_152237
|
GAS2L1
|
growth arrest-specific 2 like 1
|
|
chr1_-_36023005
|
0.865
|
NM_024874
|
KIAA0319L
|
KIAA0319-like
|
|
chr2_-_21022812
|
0.865
|
NM_021925
|
C2orf43
|
chromosome 2 open reading frame 43
|
|
chr1_+_221052845
|
0.861
|
|
HLX
|
H2.0-like homeobox
|
|
chr17_-_39968145
|
0.861
|
|
LEPREL4
|
leprecan-like 4
|
|
chr1_+_61542928
|
0.860
|
NM_001145511
|
NFIA
|
nuclear factor I/A
|
|
chr2_+_56411123
|
0.848
|
NM_001080433
|
CCDC85A
|
coiled-coil domain containing 85A
|
|
chr10_+_135483677
|
0.846
|
NM_033178
NM_012147
NM_001127387
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L7
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 7
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr1_+_100111430
|
0.841
|
NM_017734
|
PALMD
|
palmdelphin
|
|
chr19_-_58067724
|
0.838
|
NM_001039654
|
ZNF550
|
zinc finger protein 550
|
|
chr3_-_171177851
|
0.834
|
NM_001161560
NM_001161561
NM_001161562
NM_001161563
NM_001161564
NM_001161565
NM_001161566
NM_015028
|
TNIK
|
TRAF2 and NCK interacting kinase
|
|
chr11_-_116693907
|
0.834
|
NM_000482
|
APOA4
|
apolipoprotein A-IV
|
|
chr1_-_11714396
|
0.833
|
|
FBXO2
|
F-box protein 2
|
|
chr1_+_221052735
|
0.831
|
NM_021958
|
HLX
|
H2.0-like homeobox
|
|
chr4_+_126237566
|
0.830
|
NM_024582
|
FAT4
|
FAT tumor suppressor homolog 4 (Drosophila)
|
|
chr11_+_59522801
|
0.820
|
|
STX3
|
syntaxin 3
|
|
chr7_-_41742666
|
0.819
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr2_+_177134122
|
0.819
|
NM_006554
|
MTX2
|
metaxin 2
|
|
chr1_+_16091458
|
0.807
|
NM_001024216
|
FBLIM1
|
filamin binding LIM protein 1
|
|
chr1_+_61547625
|
0.806
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr2_+_177053306
|
0.802
|
NM_024501
|
HOXD1
|
homeobox D1
|
|
chr6_+_16129321
|
0.800
|
|
MYLIP
|
myosin regulatory light chain interacting protein
|
|
chr11_+_59522531
|
0.795
|
NM_001178040
NM_004177
|
STX3
|
syntaxin 3
|
|
chr19_+_56152281
|
0.793
|
NM_207115
|
ZNF580
|
zinc finger protein 580
|
|
chr4_+_100871625
|
0.792
|
|
LOC256880
|
uncharacterized LOC256880
|
|
chr4_-_102268333
|
0.791
|
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme
|
|
chr10_-_33623771
|
0.791
|
NM_001024628
NM_001024629
NM_001244972
NM_001244973
NM_003873
|
NRP1
|
neuropilin 1
|
|
chr3_+_134514044
|
0.779
|
NM_004441
|
EPHB1
|
EPH receptor B1
|
|
chr8_-_11058847
|
0.771
|
NM_173683
|
XKR6
|
XK, Kell blood group complex subunit-related family, member 6
|
|
chr1_-_179198735
|
0.770
|
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr18_+_42260774
|
0.769
|
NM_015559
|
SETBP1
|
SET binding protein 1
|
|
chr2_+_103089761
|
0.767
|
NM_001011552
|
SLC9A4
|
solute carrier family 9 (sodium/hydrogen exchanger), member 4
|
|
chr8_-_74659942
|
0.766
|
NM_001164384
|
STAU2
|
staufen, RNA binding protein, homolog 2 (Drosophila)
|
|
chr8_-_6735438
|
0.761
|
NM_005218
|
DEFB1
|
defensin, beta 1
|
|
chr14_-_88792923
|
0.755
|
|
KCNK10
|
potassium channel, subfamily K, member 10
|
|
chr19_-_36303766
|
0.746
|
|
PRODH2
|
proline dehydrogenase (oxidase) 2
|
|
chr5_-_111093030
|
0.746
|
NM_001142482
NM_001142481
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr3_+_126243161
|
0.740
|
|
CHST13
|
carbohydrate (chondroitin 4) sulfotransferase 13
|
|
chr15_-_40633122
|
0.738
|
NM_207380
|
C15orf52
|
chromosome 15 open reading frame 52
|
|
chr6_+_17600504
|
0.736
|
NM_016255
|
FAM8A1
|
family with sequence similarity 8, member A1
|
|
chr7_+_73442468
|
0.736
|
|
ELN
|
elastin
|
|
chr20_-_35885276
|
0.733
|
NM_001184731
NM_021081
|
GHRH
|
growth hormone releasing hormone
|
|
chr19_+_49376488
|
0.728
|
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr9_+_115513050
|
0.727
|
NM_001012994
|
SNX30
|
sorting nexin family member 30
|
|
chr2_-_211035913
|
0.716
|
NM_152519
|
C2orf67
|
chromosome 2 open reading frame 67
|
|
chr6_-_32163890
|
0.714
|
|
NOTCH4
|
notch 4
|
|
chr8_+_29952921
|
0.713
|
NM_001128208
NM_015344
|
LEPROTL1
|
leptin receptor overlapping transcript-like 1
|
|
chr1_+_32538497
|
0.708
|
NM_018056
|
TMEM39B
|
transmembrane protein 39B
|
|
chr17_-_71088831
|
0.703
|
|
SLC39A11
|
solute carrier family 39 (metal ion transporter), member 11
|
|
chr17_-_71088820
|
0.694
|
|
SLC39A11
|
solute carrier family 39 (metal ion transporter), member 11
|
|
chr1_+_10093149
|
0.693
|
|
UBE4B
|
ubiquitination factor E4B
|
|
chr15_+_57210972
|
0.691
|
|
TCF12
|
transcription factor 12
|